
Inflammatory factor tumor necrosis factor-α (TNF-α) activates P-glycoprotein (P-gp) by phosphorylating c-Jun and thus promotes transportation in placental cells
Introduction
Medical therapy in pregnancy under certain conditions is inevitable, especially when the mother has a chronic disease such as diabetes, depression or epilepsy (1). Drugs are frequently administered to alleviate symptoms associated with pregnancy or fight infections in mother (2). In developed countries, drug administration is quite frequent, and 27–93% of pregnant women are prescribed drugs during pregnancy (3). The high rate of prescription use during pregnancy may lead to teratogenic risk to the fetus (4).
Regardless of the specific drug administered, the degree of fetal exposure determines the risk of drug teratogenicity which is regulated, at least partially, by placental transporter proteins (5). As one of the most abundant transporter proteins, P-gp, an efflux transporter encoded by ABCB1, is thought to protect the fetus by acting as a barrier to prevent cytotoxic molecules from entering fetal circulation. P-gp is an ATP-binding cassette transporter that acts as a molecular sieve in cytotrophoblasts (CTs) and syncytiotrophoblasts (STs) in the placental barrier. The addition of a P-gp inducer drives P-gp to exert protective effects against harmful drugs (6); in contrast, the addition of the AP-1 inhibitor SR11302 causes fetal injury (7).
P-gp protects the fetus by modulating its expression and function, and its protective effects are affected by numerous exogenous and endogenous factors. As one of the most critical exogenous factors, an encompassing inflammatory environment is thought to affect P-gp both transcriptionally and post-transcriptionally (8). Tumor necrosis factor-α (TNF-α) has been reported to be primarily associated with P-gp protein levels. Hartmann et al. reported that long-term exposure to TNF-α resulted in significant upregulation of P-gp in the human liver (9). In contrast, in brain endothelial cells (BECs), which are critical components of the blood-brain barrier (BBB), long-term exposure to TNF-α significantly decreased P-gp protein levels and function (10). These contradictory results indicate that the inflammatory environment affects P-gp expression and function in the placenta.
Mammalian AP-1 belongs to the Jun (c-Jun, JunB, and JunD), Fos (c-Fos, FosB, Fra1, and Fra2), and closely related activating factor (ATF2, ATF-3, and B-ATF) families (11). Among these families, the Jun family is most abundant, and Jun family members function mainly as transcription factors by targeting many gene promoters, including the ABCB1 promoter. Tsai et al. reported that in Bewo cells, activated AP-1 transcriptionally regulates P-gp expression by binding to its promoter region (12). It has also been reported that after TNF-α exposure, activated AP-1 binds to its consensus sequence, indicating potential regulation of its downstream target genes (13). Considering these findings, we hypothesized that TNF-α may regulate P-gp indirectly by transcriptionally activating members of the AP-1 family.
Here, we sought to identify the effects of TNF-α exposure on molecule-transporting activity in placental Bewo cells. We also demonstrated that TNF-α transcriptionally activates and phosphorylates c-Jun and its functional forms and transcriptionally regulates P-gp expression and function. These findings may have clinical implications for drug use during pregnancy. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-43/rc).
Methods
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were approved by the Chengdu Women’s and Children’s Central Hospital [B2018(10)].
Cell culture and treatment
Human placental choriocarcinoma cell lines, Bewo, JEG-3 and JAR were purchased from the cell bank of the Chinese Academy of Science, and cultured in DMEM/F-12 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin at 37 °C in a humidified atmosphere of 95% air and 5% CO2. Cells were incubated with 5, 10 and 20 ng/mL recombinant tumor necrosis factor-α (rTNF-α) for 24 h and then collected for follow-up experiments.
Western blot (WB)
Cells were lysed using SoniConvert® Tissue cell sonicator (DocSense, Chengdu, China) and suspended in 200 µL of chilled lysis buffer containing 50 mM Tris-HCl (pH7.4), 150 mM NaCl, 1% Triton X-100, 5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM Na3VO4, 1 mM NaF, and 10 µM phenylmethylsulfonyl fluoride (PMSF). All the reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA). The supernatant was collected by centrifugation at 12,000 g at 4 °C for 10 min. The total protein concentration was measured using a BCA kit (Sigma-Aldrich, St. Louis, MO, USA). A 20 µg protein aliquot was fractionated on a 10% sodium dodecyl-sulfate (SDS)-polyacrylamid gel electrophoresis (PAGE) gel and transferred to a nitrocellulose membrane (Millipore, Billerica, MA, USA). The membrane was then incubated with primary antibodies at dilution of 1:1,000 at 4 °C overnight. The following primary antibodies were used (all mentioned antibodies were bought from Abcam, Cambridge, England): rabbit anti-c-Jun (Cat. No. ab40766), rabbit anti-JunB (Cat. No. ab128878), rabbit anti-JunD (Cat. No. ab181615), rabbit anti-c-Jun (S63) (Cat. No. ab32385), rabbit anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Cat. No. ab9485), rabbit anti-β-actin (Cat. No. ab8227), rabbit anti-α-Lamin B (Cat. No. ab32535), rabbit anti-ERK (Cat. No. ab32537) and rabbit anti-P-gp (Cat. No. ab234884). The membrane was then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-rabbit IgG H&L antibody, Cat. No. ab7090, Abcam) at dilution of 1:5,000 for 1 h at room temperature. The immunoassay was performed using an enhanced chemiluminescence (ECL) reagent (Cat. No. PRN2232; GE Healthcare Bio-Sciences, NJ, USA).
Flow cytometry
The cells were suspended at a final concentration of 1×106 cells/mL and washed 3 times ice-cold phosphate buffered saline (PBS). The cells were fixed in 1 mL 75% ethanol overnight at 4 °C. For the cell proliferation assay, the cells were incubated with 100 µL RNase A (Sigma-Aldrich) and 400 µL propidium iodide (PI) (Sigma-Aldrich) for 30 min at room temperature in the dark. For the apoptosis assay, cells were stained using fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA), following the manufacturer’s instructions. Labeled cells were analyzed using flow cytometry (FACS LSRII, BD Biosciences). All experiments were repeated 3 times and the results were analyzed using FlowJo 7.6 software (Tree Star, Ashland, OR, USA).
Electromobility shift assay (EMSA)
For the EMSAs, consensus probe of AP-1 was labeled with biotin. For each 20 µL containing 200 ng of total protein, 10 pM DNA probe, 2 µg of cytoplasmic protein or 2 µg of nucleus protein, 10% glycerol, 150 mM NaCl, 10 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), and 0.5 U/µL DNase inhibitor. All reagents used were bought from Sigma-Aldrich. Following incubation for 20 min at room temperature, 0.5× buffer was added to the samples and electrophoresis was performed in 6% PAGE gel (Life Technologies, Grand Island, NY, USA) at 180 V at 4 °C for 90 min. The bands were transferred onto nylon membranes (Roche, Shanghai, China) with a positive charge at 15 V at room temperature for 30 min with 0.5× TBE (Tris/Boric Acid/EDTA) as buffer and then crosslinked at 254 nm at 150 mJ/cm2. The binding bands were detected using the chemiluminescent nucleic acid detection module (Pierce, USA) following the manufacturer’s instructions.
Immunofluorescence staining
Cultured cells at 20% confluence were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 1% Triton X-100. Following blocking with 10% goat serum for 1h, cells were incubated with primary rabbit anti-P-gp antibody (Cat. No. ab234884; Abcam) at dilution of 1:100 for 2 h at room temperature. After rinsing with PBS 3 times, cells were incubated with AlexFluro secondary antibody 488-conjugated for 1 h. Then, the cells were mounted in a Vectashield with DAPI and imaged under a X71 (U-RFL-T) fluorescence microscope (Olympus, Melville, NY, USA).
Reverse transcriptional quantitative polymerase chain reaction (RT-qPCR)
Cells (5×106) were rinsed twice with ice-cold PBS and total RNA was extracted using TRIzol reagent (Life Technologies). cDNA was synthesized using a cDNA synthesis kit (Omega Bio-tek, Guangzhou, China) following the manufacturer’s instructions. RT-qPCR was conducted using SYBR Green PCR Master Mix (Life Technologies) following the manufacturer’s instructions. The following primers were used: (I) c-Jun: forward, 5'-TCCAAGTGCCGAAAAAGGAAG-3' and reverse, 5'-CGAGTTCTGAGCTTTCAAGGT-3'; (II) ABCB1: forward, 5'-TTGCTGCTTACATTCAGGTTTCA-3' and reverse, 5'-AGCCTATCTCCTGTCGCATTA-3'; (III) β-actin: forward, 5'-CATGTACGTTGCTATCCAGGC-3' and reverse, 5'-CTCCTTAATGTCACGCACGAT-3'. Thermal cycling was carried out for 10 min at 95 °C followed by 35 cycles of 15 s at 95 °C and 1 min at 60 °C. Experiments were performed in triplicate.
Chromatin immunoprecipitation (ChIP)
Cells (5×106) were collected, suspended in 1 mL of PBS, and incubated in 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. A Misonix 3000 sonicator (Cole-Parmer Instrument Company, Vernon Hills, IL, USA) with microtip was employed to generate 200–500 bp DNA fragments according to the following protocol: power 7 (approximately 7 W), 24 cycles (30 s sonication, 90 s rest). ChIP was performed as follows: 100 µL of sonicated cell lysate was incubated with 10 µg of c-Jun antibody (Cat. No. ab40766; Abcam) or rabbit IgG (Sigma-Aldrich) for 2 h at room temperature. Then, 20 µL of protein A agarose beads (Life Technologies) was added to each reaction for 1 h incubation at room temperature. The beads were washed with 1 mL chilled PBS 3 times and eluted using elution buffer containing 1% SDS and 100 mM NaHCO3. DNA fragments were purified from eluant using Cycle Pure Kit (Omega Bio-tek) and 5 µL of purified DNA fragment was used as template for PCR detection. Primers were used for PCR amplification of the obtained DNA in the enrichment multiple assay of c-Jun protein-binding ABCB1 promoter or dihydrofolate reductase (DHFR) 5' untranslated region (5'UTR). The following primers were used: (I) P1: forward, 5'-AAATGGCTACATGAGAGCGG-3'; reverse, 5'-TCTTTGCTCCACTGACATGAAA-3'; (II) P2: forward, 5'-GCAAGGGGACCAGGTAGGTTTC-3'; reverse, 5'-TCAGGTAAAGATCTCTATCAAA-3'; (III) P3: forward, 5'-CAGGAAATCTGAAAGCCTGAC-3'; reverse, 5'-CGGTTTCTAGAGAGGTGCA-3'. PCR was performed using SYBR Green PCR Master Mix (Life Technologies) on an Applied Biosystems 7500 real-time system (ABI 7500HT instrument). Thermal cycling was performed out for 5 min at 50 °C, 5 min at 95 °C and 35 cycles of 30 s at 95 °C and 60 s at 60 °C. Experiments were performed in triplicates.
Luciferase activity assay
pGL3 containing the ABCB1 consensus-binding sequence (pGL3-ABCB1-BS, upper: 5'-TAAGTATGACTCACCAGGGAC-3') firefly luciferase reporter plasmids were used in conjunction with the control pGL3-basic vector (Promega, Madison, WI, USA) and internal control plasmid pRL-SV40 (Promega, Madison, WI, USA). Cells (5×105) were cultured in 6-well plates for attachment and transfected with the indicated plasmid using LipofectamineTM 2000 (Life Technologies) according to the manufacturer’s instructions. Cells were collected 48 h later. The flame and luciferase activities of the cell lysates were analyzed using dual a luciferase reporter assay system (Promega, Madison, WI, USA). Data are represented as fold-induced comparisons with pGL3-BASIC vectors. The entire experiment was independently repeated 3 times.
Uptake of DiOC2(3) and Rh123
Rhodamine 123 (Rh123) and 3,3'-diethyloxacarbocyanine iodide [DiOC2(3)] were used to identify the functional efflux activity of P-gp in cells. Briefly, 2×106 Bewo cells were harvested and incubated with 10 µg/mL Rh123 for 90 min or 1 µg/mL DiOC2(3) for 20 min. After incubation, the cells were washed with cold PBS 3 times and each aliquot of cells was incubated with 20 µM vinblastine in dye-free solution at 37 °C for 60 min. Ice-cold PBS was used to terminate the efflux. The fluorescence of Rh123 and DiOC2(3) was quantified using a microplate spectrophotometer (Synergy 2 Multi-Mode Microplate Reader; BioTek, Winooski, VT, USA) at the excitation and emission wavelengths of 485 and 530 nm, respectively.
Detection of efflux ratio of digoxin
The cells were cultured overnight in 24-well plates (BD-Corning, Corning, NY, USA), with each well containing 1×105 cells. After three washes with PBS, 0.5 mL Hanks’ balanced salt solution (HBSS) supplemented with 0.1 µM [3H]digoxin, Tween 80 (0.1% and 1%), and P85 (0.1% and 1%) was added to initiate digoxin accumulation. After washing with ice-cold HBSS, cells were lysed with 0.1 mL 10% SDS, stabilized with a cocktail solution (Perkin Elmer Inc.; Boston, MA, USA), and incubated overnight. The radioactivity of [3H]digoxin in the cell lysate was measured using a Microbeta 2 liquid scintillation counter (PerkinElmer Inc., Boston, MA, USA).
Statistical analysis
Statistical analyses were performed using SPSS version 13.0. Data are expressed as mean ± standard deviation (SD). The statistical significance of the difference between groups was assessed using Student’s t-test and one-way analysis of variance followed by Bonferroni post-hoc analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of Jun family members and their regulation by rTNF-α in Bewo cells
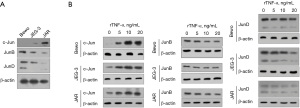
The protein expression of c-Jun, JunB, and JunD in Bewo, JEG-3, and JAR cells was analyzed using WB. As shown in Figure 1A, c-Jun, JunB, and JunD were differentially expressed in all the three cell lines. Lower levels of c-Jun were observed in the Bewo and JEG-3 cells than in the JAR cells. In contrast, JunD protein expression was higher in Bewo cells than that in JEG-3 and JAR cells. To investigate the expression of Jun family members, namely c-Jun, JunB, and JunD, after rTNF-α treatment, we first exposed Bewo cells to 5, 10, or 20 ng/mL rTNF-α and analyzed the protein levels of Jun family members using WB (Figure 1B). c-Jun protein was clearly regulated by the addition of rTNF-α in a dose-dependent manner. c-Jun expression was significantly upregulated in all three cell lines. Interestingly, rTNF-α significantly upregulated c-Jun in JEG-3 cells but did not affect JunB protein levels. rTNF-α was also remarkably downregulated of JunD protein in JAR cells, but not in Bewo or JEG-3 cells. These results suggest that rTNF-α may exert its function by regulating c-Jun protein, but not JunB or JunD. As shown in Figure 2A, c-Jun was the most strongly affected AP-1 family member.
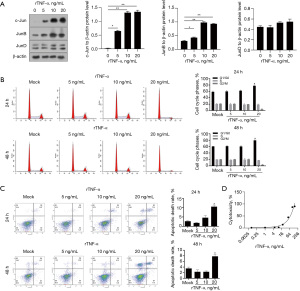
We then used Bewo cells to analyze whether rTNF-α affects proliferation and/or apoptosis. As shown in Figure 2B, 20 ng/mL rTNF-α significantly blocked the cell phase at G1/G0 after 24 h of treatment, and 5 or 10 ng/mL rTNF-α negligibly affected cell viability. Analysis of cells in different cell cycle phases consistently showed that 20 ng/mL rTNF-α, but not 5 or 10 ng/mL rTNF-α, significantly increased the percentage of cells in G1/G0 (Figure 2C). After 48 h of rTNF-α treatment, 20 ng/mL rTNF-α significantly increased the apoptotic cell death rate, but neither 5 nor 10 ng/mL rTNF-α had this effect. To determine whether cytotoxicity was induced by rTNF-α in Bewo cells, a crystal violet incorporation assay was performed after treatment of Bewo cells with different concentrations of rTNF-α. As shown in Figure 2D, 10 ng/mL rTNF-α caused no detectable cytotoxicity in the Bewo cells. We also treated JEG-3 cells with 10 ng/mL rTNF-α for 24 and 48 h and then performed cell cycle and apoptosis analyses. As expected, treatment with 10 ng/mL rTNF-α negligibly affected the cell cycle distribution and apoptosis of JEG-3 cells (Figure S1). These data suggested that 5 and 10 ng/mL rTNF-α significantly affected c-Jun protein levels without obviously affecting cell proliferation or apoptosis, indicating its potential effect on other physiological processes.
rTNF-α enhanced phosphorylation of c-Jun and promoted its DNA binding activity
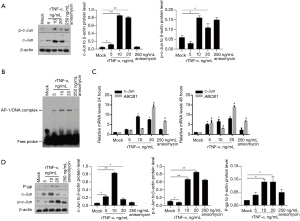
After 24 h of rTNF-α treatment, c-Jun phosphorylation was analyzed in Bewo cells. After the addition of 250 ng/mL anisomycin, a c-Jun activator, the phosphorylation of c-Jun increased without affecting the c-Jun protein (Figure 3A). rTNF-α treatment significantly increased the expression of c-Jun and p-c-Jun (Figure 3A). It has been reported that rTNF-α activates AP-1 transcriptional activity in several cell types (14), which prompted us to test whether rTNF-α activates AP-1 transcriptional activity in Bewo cells. The results demonstrated that the addition of rTNF-α promoted the binding of the AP-1 consensus probe in cell lysates, indicating the potential activation of AP-1 by rTNF-α treatment (Figure 3B). Considering that activation of AP-1 may transcriptionally activate ABCB1 (15), we performed quantitative PCR to measure ABCB1 mRNA levels under different concentrations of rTNF-α for 24 and 48 h. As shown in Figure 3C, both rTNF-α and anisomycin treatment upregulated ABCB1 mRNA at 24 and 48 h time points. We also detected P-gp protein levels after rTNF-α treatment and found that rTNF-α treatment significantly increased P-gp protein levels (Figure 3D).
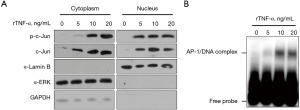
rTNF-α treatment increased total c-Jun and phosphorylated c-Jun (p-c-Jun) in both cytoplasm and nucleus
The results showed that rTNF-α promoted the transcriptional activity of ABCB1 in Bewo cells. Therefore, we sought to determine whether rTNF-α could induce increase in p-c-Jun in both the cytoplasm and nucleus. As shown in Figure 4A, we obtained the cytoplasmic portion without α-Lamin B contamination and the nuclear portion without α-extracellular-signal-regulated kinase (ERK) or GAPDH contamination. In both the cytoplasm and nucleus, rTNF-α obviously increased c-Jun and p-c-Jun levels in a concentration-dependent manner. To confirm the activation of p-c-Jun in the nucleus, we extracted nuclear lysates from Bewo cells after 24 h of treatment with 10 ng/mL rTNF-α and detected its binding to AP-1 consensus DNA cis-elements. By performing EMSA, co-incubation of nuclear fraction with AP-1 consensus DNA resulted in a shifted band (Figure 4B). Considering that rTNF-α treatment obviously increased p-c-Jun in the nuclear fraction, the shifted band was potentially composed of p-c-Jun and AP-1 consensus DNA probe.
P-c-Jun by rTNF-α treatment binds specifically to ABCB1 gene promoter region
To identify and confirm the regulation of ABCB1 expression by p-c-Jun, we predicted putative p-c-Jun binding sites on the ABCB1 promoter region using the online bioinformatics tool PROMO ALGGEN (TRANSFAC 8.1) and found one putative binding site located −2,000 bp upstream. After a ChIP assay, the IP products were detected using three pairs of primers: two containing putative binding sites (−2,200 to −1,900 bp and −2,100 to −1,800 bp) and one considered as the negative control (−200 to 200 bp). As illustrated in Figure 5A, significant enrichment of DNA fragments was observed by using two pairs of primers containing putative binding sites after incubation with rTNF-α for 24 h SP600125 (1.75 µM), a c-Jun phosphorylation inhibitor, decreased the fold enrichment, indicating that the binding of c-Jun to the ABCB1 promoter was, at least partially, dependent on the phosphorylation of c-Jun. To confirm the binding of p-c-Jun to the ABCB1 promoter fragment, a fragment from −2,200 to −1,900 bp in the ABCB1 promoter was inserted before the luciferase promoter. As shown in Figure 5B, rTNF-α treatment decreased luciferase activity by stimulating the binding of transcription factors to the inserted fragments, which was reversed by the inhibition of c-Jun phosphorylation. Consequently, rTNF-α promoted the presence of P-gp on the surface membrane of Bewo cells, which was reversed by 1.75 µM SP600125 (Figure 5C). ChIP was performed on rTNF-α-treated JEG-3 cells to confirm the effect of rTNF-α on the binding of c-Jun to the ABCB1 promoter region. As expected, c-Jun binding to the ABCB1 promoter region increased (at three sites: −2,200 to −1,900 bp, −2100 to −1,800 bp, and −200 to +200 bp) (Figure S2A). rTNF-α treatment also decreased luciferase activity, potentially by stimulating c-Jun phosphorylation (Figure S2B).
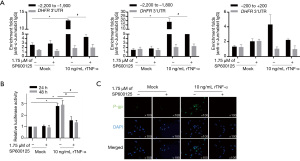
rTNF-α-stimulated c-Jun regulates P-gp function
To determine the regulatory effect of c-Jun on P-gp function, two P-gp-specific substrates, Rh123 and DiOC2(3), were used. First, SP600125 was administered to inhibit the phosphorylation of c-Jun and then SR11302 was added to inhibit the activity of AP-1. As shown in Figure 6A, SP600125 and SR11302 significantly affected c-Jun protein levels, and the addition of SR11302, but not SP600125, inhibited c-Jun phosphorylation. The uptake of Rh123 and DiOC2(3) was assessed, and the results showed that both Rh123 and DiOC2(3) were decreased by the addition of vinblastine, a P-gp competitive inhibitor (Figure 6B). We then evaluated the effect of c-Jun, AP-1 and P-gp on the transport of digoxin across Bewo cell monolayers in both the apical (AP) to basolateral (BL) (AP-BL) and BL to AP (BL-AP) directions. As illustrated in Figure 6C, the efflux ratio was significantly increased by rTNF-α and significantly decreased by vinblastine. These data indicate that c-Jun is critical for regulating P-gp protein expression and function.
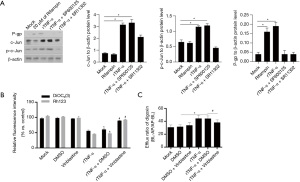
Discussion
P-gp is an abundant transporter in the placenta and exerts protective effects against cytotoxic agents during pregnancy. The induction and inhibition of P-gp function have been reported to tightly regulate the efflux rate of a specific P-gp substrate. TNF-α has been reported to affect P-gp protein expression, despite inconsistent findings (9,10). However, little is known about the exact effect of TNF-α on P-gp in the placenta. Previous studies have revealed that AP-1 expression and function are regulated by multiple mechanisms, including phosphorylation by various mitogen-activated protein kinases (MAPKs) (16) and the influence of inflammatory factors, particularly TNF-α (12,17). In this study, we found that rTNF-α induced upregulation of P-gp expression and binding activity to the AP-1 consensus DNA binding sequence, while the effects were attenuated by pretreatment with inhibitors of AP-1 (SR11302) or P-gp (SP600125). rTNF-α treatment promoted P-gp transport activity by upregulating c-Jun and p-c-Jun. These results demonstrate that rTNF-α may be a critical regulator of the barrier that prevents cytotoxic molecules from entering the fetal circulation.
Accumulating evidence demonstrates that in cancers, TNF-α stimulates c-Jun through the activation of c-Jun N-terminal kinase (JNK), which is a classical signaling pathway tightly associated with inflammation (18). In this study, we analyzed c-Jun mRNA and protein levels in response to rTNF-α treatment and found that rTNF-α induced c-Jun expression and phosphorylation in a dose- and time-dependent manner in Bewo cells, consistent with previous reports demonstrating that TNF-α facilitates a variety of pathophysiological activities through transcriptional and posttranscriptional regulation of c-Jun expression (19,20). Considering that TNF-α increases both c-Jun and p-c-Jun levels in a dose- and time-dependent manner, it is still unclear whether TNF-α directly promotes c-Jun phosphorylation or not.
TNF-α primarily interacts with TNF receptor 1 (TNFR1) and/or TNF receptor 2 (TNFR2) and thus mediates a variety of cell signaling processes associated with inflammatory induction (21,22). Kobelt et al. reported that in colorectal cancer, long-term exposure of cancer cells to TNF-α upregulated c-Jun by interacting with TNFR1 but not TNFR2 (19), indicating that in Bewo cells, TNF-α may also function depending on the presence of TNFR1. In future investigations, it will be worth determining whether TNF-α regulates c-Jun or AP-1 by interacting with TNFR1.
The c-Jun protein and p-c-Jun protein induced by TNF-α share their main functional portions with the transcription factor AP-1, which was previously reported to transcriptionally drive ABCB1 gene expression (12,13). Using the online bioinformatics tool PROMO ALGGEN (TRANSFAC 8.1), the predicted binding site of c-Jun to the core promoter region of ABCB1 was identified near the nucleotide −2,000 bp from the transcriptional start site. In the mock group (without TNF-α pretreatment) (Figure 5A), a 4-h pretreatment with 1.75 µM SP600125, the inhibitor of c-Jun phosphorylation, failed to affect the fold enrichment of c-Jun in the promoter region of the ABCB1 gene, indicating that endogenous p-c-Jun has no obvious function in Bewo cells. This finding further demonstrated that TNF-α treatment significantly enhanced c-Jun function as a transcription factor, especially when targeting ABCB1.
Mammalian AP-1 proteins are homologous and heterogeneous and are composed of the Jun (c-Jun, JunB, and JunD), Fos (c-Fos, FosB, Fra1, and Fra2), and closely related activating factor (ATF-2, ATF-3, and B-ATF) families (11). AP-1 proteins are critically involved in the regulation of physiological processes, including embryonic development and cell survival or death, by inducing and/or repressing tumor suppressor genes (23,24). In three placental cell lines, Bewo, JEG-3 and JAR, TNF-α administration consistently regulated the c-Jun protein but did not affect other Jun family members, prompting us to focus on c-Jun and its phosphorylated form. c-Jun is reported to be one of the most important members of the Jun family. For instance, c-Jun is required for the proliferation of fibroblasts and other nontumor cultured cells (25,26). In tumor cells, c-Jun protects cells from p53-induced apoptosis following ultraviolet irradiation-induced DNA damage (27).
Conclusions
In summary, we concluded that rTNF-α activates P-gp by directly upregulating c-Jun expression and phosphorylation in Bewo cells. A complete understanding of the regulatory roles of TNF-α is pivotal and may enable the development of molecules that preserve the beneficial effects of TNF-α and new strategies for administration to protect the fetus from the adverse effects of maternal pharmacology.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant Nos. 8217060614 and 8210060921), the Sichuan Science and Technology Program (grant Nos. 2018JY0355 and 2021YJ0190), Medical Technology Program of Health Commission of Sichuan Province (grant No. 21PJ129) and Outstanding Talents Program of Chengdu Women’s and Children’s Central Hospital (grant No. YC2021001).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-43/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-43/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-43/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-43/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were approved by the Chengdu Women’s and Children’s Central Hospital [B2018(10)].
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Daud AN, Bergman JE, Bakker MK, et al. P-Glycoprotein-Mediated Drug Interactions in Pregnancy and Changes in the Risk of Congenital Anomalies: A Case-Reference Study. Drug Saf 2015;38:651-9. [Crossref] [PubMed]
- Bakker MK, Jentink J, Vroom F, et al. Drug prescription patterns before, during and after pregnancy for chronic, occasional and pregnancy-related drugs in the Netherlands. BJOG 2006;113:559-68. [Crossref] [PubMed]
- Daw JR, Hanley GE, Greyson DL, et al. Prescription drug use during pregnancy in developed countries: a systematic review. Pharmacoepidemiol Drug Saf 2011;20:895-902. [Crossref] [PubMed]
- Andrade SE, Gurwitz JH, Davis RL, et al. Prescription drug use in pregnancy. Am J Obstet Gynecol 2004;191:398-407. [Crossref] [PubMed]
- Staud F, Cerveny L, Ceckova M. Pharmacotherapy in pregnancy; effect of ABC and SLC transporters on drug transport across the placenta and fetal drug exposure. J Drug Target 2012;20:736-63. [Crossref] [PubMed]
- Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev 2003;55:3-29. [Crossref] [PubMed]
- Zhou SF. Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica 2008;38:802-32. [Crossref] [PubMed]
- Cario E. P-glycoprotein multidrug transporter in inflammatory bowel diseases: More questions than answers. World J Gastroenterol 2017;23:1513-20. [Crossref] [PubMed]
- Hartmann G, Kim H, Piquette-Miller M. Regulation of the hepatic multidrug resistance gene expression by endotoxin and inflammatory cytokines in mice. Int Immunopharmacol 2001;1:189-99. [Crossref] [PubMed]
- Poller B, Drewe J, Krähenbühl S, et al. Regulation of BCRP (ABCG2) and P-glycoprotein (ABCB1) by cytokines in a model of the human blood-brain barrier. Cell Mol Neurobiol 2010;30:63-70. [Crossref] [PubMed]
- Subramanian D, Bunjobpol W, Sabapathy K. Interplay between TAp73 Protein and Selected Activator Protein-1 (AP-1) Family Members Promotes AP-1 Target Gene Activation and Cellular Growth. J Biol Chem 2015;290:18636-49. [Crossref] [PubMed]
- Tsai YT, Lozanski G, Lehman A, et al. BRAFV600E induces ABCB1/P-glycoprotein expression and drug resistance in B-cells via AP-1 activation. Leuk Res 2015;S0145-2126(15)30371-4. Epub ahead of print. [Crossref] [PubMed]
- Kankaanranta H, Ilmarinen P, Zhang X, et al. Tumour necrosis factor-α regulates human eosinophil apoptosis via ligation of TNF-receptor 1 and balance between NF-κB and AP-1. PLoS One 2014;9:e90298. [Crossref] [PubMed]
- Keystone EC, Ware CF. Tumor necrosis factor and anti-tumor necrosis factor therapies. J Rheumatol Suppl 2010;85:27-39. [Crossref] [PubMed]
- Roy KR, Reddy GV, Maitreyi L, et al. Celecoxib inhibits MDR1 expression through COX-2-dependent mechanism in human hepatocellular carcinoma (HepG2) cell line. Cancer Chemother Pharmacol 2010;65:903-11. [Crossref] [PubMed]
- Shaulian E. AP-1--The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal 2010;22:894-9. [Crossref] [PubMed]
- Kim H, Yang WS, Htwe KM, et al. Dipterocarpus tuberculatus Roxb. Ethanol Extract Has Anti-Inflammatory and Hepatoprotective Effects In Vitro and In Vivo by Targeting the IRAK1/AP-1 Pathway. Molecules 2021;26:2529. [Crossref] [PubMed]
- Yi M, Wang D, Chen Y, et al. β-Asarone suppresses TNF-α expression through DNA methylation and c-Jun-mediated transcription modulation in scratch-injured neuronal cells. J Biochem Mol Toxicol 2021;35:e22798. [Crossref] [PubMed]
- Kobelt D, Zhang C, Clayton-Lucey IA, et al. Pro-inflammatory TNF-α and IFN-γ Promote Tumor Growth and Metastasis via Induction of MACC1. Front Immunol 2020;11:980. [Crossref] [PubMed]
- Wang J, Tai G. Role of C-Jun N-terminal Kinase in Hepatocellular Carcinoma Development. Target Oncol 2016;11:723-38. [Crossref] [PubMed]
- Chu WM. Tumor necrosis factor. Cancer Lett 2013;328:222-5. [Crossref] [PubMed]
- McDermott MF. TNF and TNFR biology in health and disease. Cell Mol Biol (Noisy-le-grand) 2001;47:619-35. [PubMed]
- Jochum W, Passegué E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene 2001;20:2401-12. [Crossref] [PubMed]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol 2002;4:E131-6. [Crossref] [PubMed]
- Niu YN, Wang K, Jin S, et al. The intriguing role of fibroblasts and c-Jun in the chemopreventive and therapeutic effect of finasteride on xenograft models of prostate cancer. Asian J Androl 2016;18:913-9. [PubMed]
- Hammouda MB, Ford AE, Liu Y, et al. The JNK Signaling Pathway in Inflammatory Skin Disorders and Cancer. Cells 2020;9:857. [Crossref] [PubMed]
- Shaulian E, Schreiber M, Piu F, et al. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell 2000;103:897-907. [Crossref] [PubMed]


