
Functional characterization of a KCNAB3 genetic epilepsy with febrile seizures plus adult mouse model
Introduction
Genetic epilepsy with febrile seizures plus (GEFS+), previously known as generalized epilepsy with febrile seizures (FS) plus, was first reported in a large Australian family by Scheffer and Berkovic (1) in 1997. Later, focal epilepsy phenotypes, such as temporal lobe and frontal lobe epilepsy, were observed in GEFS+ families. Thus, Scheffer and Berkovic (2) proposed the current denomination of GEFS+. In 2001, this disease was identified as a new epileptic syndrome by the International League Against Epilepsy and is characterized by phenotypic and genetic heterogeneity. GEFS+ is a genetic epilepsy syndrome with a broad phenotypic spectrum (3). Different members of the same family may have phenotypes ranging from slight FS to serious epileptic encephalopathy, such as Dravet syndrome (4-7). The etiopathology of GEFS+ is complex, and there is strong evidence that it has a genetic predisposition (8). GEFS+ is a complex autosomal dominant disorder (9). According to family analysis, most GEFS+ can be attributed to autosomal dominant inheritance with incomplete dominance, the GEFS+ phenotypic penetrance has been reported to range from 62–76%, a study has found double inheritance in GEFS+ families, GEFS+ may have complex inheritance modes, such as polygenic inheritance (10). Mutations in a variety of pathogenic genes are involved in the pathogenesis of GEFS+, usually caused by mutations in gene encoding voltage-gated sodium channel protein subunit SCN1A (9). SCN2A, SCN1B, GABRG2, and GABRD are also common pathogenic genes associated with GEFS+. Other genes such as SCN9A, STX1B, and Fgf13 may also be involved in the pathogenesis of GEFS+.
Previously, we identified a new missense mutation in the KCNAB3 gene H258R (HR) in a GEFS+ family. This mutation was first reported by our team. There are 18 members in this family, 4 were affected with GEFS+. Whole-exome sequencing was performed to assess for genetic mutations. KCNAB3HR was shared by three affected and one unaffected family member. This mutation was thought to affect protein function by the bioinformatics tools. For patients with genetic epilepsy, the connection between a specific mutation and their behavioral symptoms is largely unknown (11). To clarify the relationship between the KCNAB3 gene mutation (H258R) and GEFS+, wild and mutant plasmids were constructed and transfected into human embryonic kidney 293 (HEK293) cells for patch-clamp detection. The whole cell patch-clamp detection results suggested that this mutation caused the inactivation of potassium channels, reduced potassium outflow, increased cell excitability, and promoted convulsion (12). In the present study, to examine the effects of this mutation on potassium channels in the mammalian brain, we introduced the H258R mutation into the mouse KCNAB3 gene using CRISPR/Cas9. A patch clamp was then used to detect potassium currents in the pyramidal cells of the hippocampal CA1 region. Hippocampal sclerosis (HS) is considered to be an important pathological change of epilepsy, especially temporal lobe epilepsy (TLE). Many TLE patients with HS have a history of FS in childhood. The clinical manifestations of GEFS+ are mainly heat-related convulsions, therefore, hippocampal cells were selected as the research objects in this study (13). A protocol was prepared before the study without registration. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-436/rc).
Methods
Generation of KCNAB3HR mice
CRISPR/Cas9 technology was used to knock-in the human KCNAB3 exon 10 (H258R) expression frame at exon 10 of the mouse KCNAB3 gene through homologous recombination. Briefly, Cas9 messenger RNA (mRNA) and guide RNA (gRNA) were obtained by in-vitro transcription. In-fusion cloning was used to construct a donor vector comprising 3.0 kb 5'homologous arms, human KCNAB3 exon10 (H258R) and 3.0 kb 3'homologous arms. Cas9 mRNA, gRNA, and the donor vector were microinjected into the fertilized eggs of C57BL/6J mice (Bought from Shanghai Model Organisms Center, Inc., Shanghai, China) to obtain the F0 generation mice. The mice confirmed to be positive by polymerase chain reaction (PCR) amplification and sequencing were mated with C57BL/6J mice to obtain the F1 generation.
Animal experiments were performed under a project license (No. KY-D-2021-120-06) granted by the ethics committee of Guangdong Provincial People’s Hospital, in compliance with Guangdong Provincial People’s Hospital guidelines for the care and use of animals.
PCR
To screen for mice with the KCNAB3HR mutation, the DNA was amplified using a KCNAB3-specific forward primer and a reverse primer. PCR amplification was performed for 35 cycles at 98 ℃ for 15 s, 61 ℃ for 15 s, and 68 ℃ for 30 min. The reaction mixture consisted of 13.2 µL of ddH2O, 2 µL of GXL PCR buffer, 2 µL of 2.5-mM dNTP, 0.8 µL of GXL DNA polymerase, 0.5 µL of forward primer, 0.5 µL of reverse primer, and 1 µL of genomic DNA.
Cell separation
The hippocampal CA1 region was dissected from brain slices from the experimental mice and placed in protease [Hanks’ Balanced Salt Solution (HBSS) configuration] for digestion at 33.3 ℃ for 30 min. The whole digestion process was treated with high purity oxygen. After digestion, 3 mL of oxygen-saturated HBSS Na+ solution was added to each tube of enzyme solution to terminate the protease action. This was repeated 3 times; then, 2 mL of oxygen-saturated low Ca2+ N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid (HEPES) solution was added. The tissue was sequentially agitated with large, medium, and small caliber Pasteur pipettes and left to stand for 2 min. After agitation with the large and medium caliber pipettes, the supernatant was removed and placed in a new 15-mL centrifuge tube. Next, the tissue was agitated with a small caliber Pasteur pipette and left to stand for 2 min. The supernatant was then sucked out and mixed with the 1st supernatant. Mixed cell suspensions were inoculated in a 3.5-cm cell culture dish treated with polylysine at 2 mL per dish. After standing for 10 min, the cells had adhered to the wall and were ready for the patch clamp.
Patch clamp
A capillary glass tube (BF150-86-10, Sutter Instruments) was drawn into the recording electrode with a microelectrode drawing instrument (P97, Sutter Instruments). The microelectrode manipulator (MP225, Butter Instrument) was operated under an inverted microscope (MF53, micro-shot). The recording electrode contacted the cell, and negative pressure was pumped to form a GΩ seal. After achieving the GΩ seal, rapid capacitive compensation was performed. Continuous negative pressure served to absorb the membrane and achieve the whole cell recording mode. Next, the slow capacitance was compensated, and the membrane capacitance and series resistance were recorded. The experimental data were collected using an EPC-10 amplifier (HEKA) and stored with PatchMaster (HEKA) software. All the experiments were performed at room temperature. The following fluids were used for the recording: extracellular fluid: 140 mM of NaCl, 3.5 mM of KCl, 2 mM of CaCl2·2H2O, 1 mM of MgCl2·6H2O, 10 mM of HEPES, 10 mM of D-glucose, and 1.25 mM of NaH2PO4, with a NaOH adjusted pH of 7.4. The following intracellular fluids were used: 20 mM of KCl, 115 mM of K-aspartic, 1 mM of MgCl2·6H2O, 5 mM of EGTA, 10 mM of HEPES, and 2 mM of Na2-ATP, with a KOH adjusted pH of 7.2.
Statistical analysis
After exporting the current diagram recorded by PatchMaster, the original current data were output by Igor Pro software and saved in Excel. The current density was calculated using the I-V curve, and a collaboration diagram was then prepared with GraphPad Prism 8.0 software, and the current obtained under the maximum activated voltage was subsequently compared. The t-test was used for the statistical analysis, and a P value <0.05 was considered statistically significant.
Results
Generation of KCNAB3HR mutant mice
The injected fertilized eggs were transplanted into pseudo-pregnant female mice. The F0-generation mice were born at about 20 days and identified by PCR amplification. Positive 5' arm homologous recombination with 5'CTGACCTCATCAACGTGCCT'3 as the forward primer and 5'CGCTCCGCTTGTTCACACAC'3 as the reverse primer resulted in an amplification that produced a 3.4 kb fragment, while positive 3' arm homologous recombination with 5'GAGCGCCATCTGTTTCAGA'3 as the forward primer and 5'CTGGCCTTTTGCTGGTTTCC'3 as the reverse primer resulted in an amplification that produced a 3.2 kb fragment (see Figure 1). Due to the rapid cleavage rate of the fertilized eggs in the early stages, the obtained F0-generation mice were chimeras, which may not have a stable inheritance. Thus, the passage was required to obtain stable inheriting F1 mouse generations.
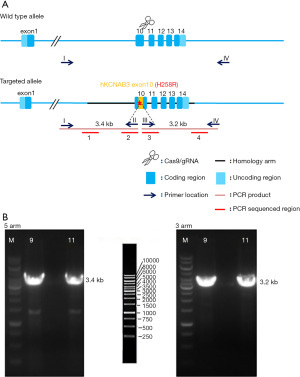
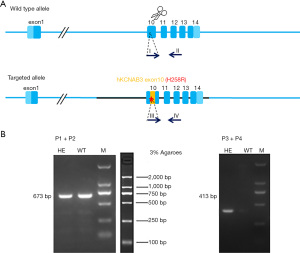
The F0-generation positive mice were mated with wild-type (WT) C57BL/6J mice to obtain F1-generation mice. The F1-generation mice were identified by PCR and sequencing; 4 sequencing reactions were performed (see Figure 2). The F1-generation mice genotypes were amplified using the primer pairs (P1: 5'GAGCAAGCGGAGAACCACT'3, P2: 5'AAACAGAGCTGACACCCTCG'3) and (P3: 5'GCGGAGCGCCATCTGTTTCA'3, P4: 5'TGGCTTGTTGCTTCTTGCCT'3). The genotypes were as follows: WT, only (P1, P2) amplified 673 bp band, (P3, P4) no band; heterozygous type: (P1, P2) amplified 673 bp band and (P3, P4) amplified 413 bp band; homozygous type: (P1, P2) no band, (P3, P4) amplified 413 bp band (see Figure 3).
Potassium current of CA1 pyramidal neurons in KCNAB3HR mice
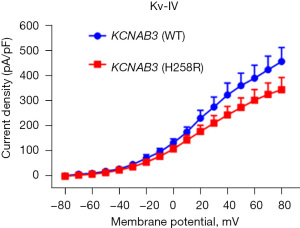
A patch clamp was used to detect the total potassium current of the pyramidal cells (n=10) in the hippocampal CA1 region of the KCNAB3 (WT) and KCNAB3 (H258R) adult mice; changes in the current were recorded as voltage increased. The current stimulus programme was as follows: after forming the whole cell seal, the cell membrane voltage was clamped at −80 mV and maintained for 50 ms. The cell membrane voltage was stimulated from −80 mV to +80 mV with 10 mV steps and maintained for 200 ms. Finally, it was recovered to −80 mV and maintained for 50 ms. The original current diagram showed that the total potassium current of the H258R group decreased as voltage increased (see Figure 4).
We plotted an I-V curve and compared the current difference at the maximum voltage. The result showed that the total potassium current decreased in the H258R group, but there were no significant differences in the current at the maximum voltage (+80 mV) (P>0.05; see Figure 5).
Discussion
At present, the genetic research of GEFS+ mainly focuses on pathogenic genes. The pathogenic genes related to GEFS+ are mainly divided into two categories: (I) genes encoding voltage-gated sodium channel protein subunits (SCN1A, SCN2A, SCN1B, SCN9A); (II) genes encoding ligand-gated chloride channel GABAA subunits (GABRG2, GABRD). Voltage-gated sodium channels (Nav) exist in the membrane of most excitable tissues and play an important role in the generation and propagation of action potentials. GABA is the most important inhibitory transmitter in the central nervous system. It is mainly located in the postsynaptic membrane of neurons in the central nervous system. It inhibits the excitability of neurons and is the target of neuroexcitatory inhibitory drugs such as benzodiazepines, barbiturates, and steroids. Others include STX1B, which encodes synapse fusion protein 1B, and Fgf13, which encodes fibroblast growth factor protein.
The potassium channel is the most complex ion channel and has the most known subtypes. It is widely expressed in the body and plays a role in maintaining neuron membrane potential, regulating excitability, and participating in cardiac myocyte repolarization. Further, it is associated with neurological and cardiac diseases. Recently, several cases of GEFS+ caused by mutations in potassium channel genes have been reported. Notably, Zou et al. (14) reported the heterozygous missense mutation P537S of KCNQ4 in a GEFS+ family. Yu et al. (15) found a KCNQ3 mutation (c.2128T > C) related to GEFS+. Tian et al. (16) summarized the clinical characteristics of a GEFS+ family caused by a KCNT2 gene mutation and theorized that GEFS+ induced by this mutation may be insensitive to antiepileptic drugs. In a previous study, we identified the heterozygous missense mutation p.H258R of the KCNAB3 gene encoding the voltage-gated potassium channel β3 subunit (Kvβ3) in a GEFS+ family.
Kvβ3 is primarily expressed in the rat brain (17). Human Kvβ3 mRNA is expressed in the brain and predominantly detected in the cerebellum (18). Kvβ3 subunits have an N-terminal inactivating domain that confers rapid N-type inactivation to Kv channels (19). The co-expression of rat Kvβ3 in Xenopus oocytes leads to the rapid inactivation of Kv1.4 channels but not of other Kv1α channels (17). However, the co-expression of Kv1.5 and Kvβ3 in a mammalian cell line yields rapidly inactivating A-type channels (18). In contrast to findings on the Xenopus oocyte expression system, Kvβ3 confers rapid inactivation to all Kv1 channels except Kv1.3 channels in Chinese hamster ovary cells (20).
In previous studies, HEK293 cells expressing human Kv1.1 and Kvβ3 were detected by the patch clamp. The present results showed that the current of cells expressing only Kv1.1 exhibited no inactivation, while the current of cells co-expressing Kvβ3 was inactivated; the inactivation characteristics were more obvious in cells co-expressing Kvβ3 (H258R). Together, these findings indicate that the Kvβ3 subunit promotes potassium channel inactivation and that the H258R mutation further accelerates this inactivation. In the present study, we induced the expression of the human KCNAB3 gene in mice using a knock-in technique and then used a patch clamp to detect potassium currents in the pyramidal cells in the hippocampal CA1 region of the mouse brain. Our results showed that the total potassium current decreased in the H258R group but was not significantly different from the current at the maximum voltage (+80 mV). This may be explained by the different inactivation activity of Kvβ3 in various expression systems. Compared to cell-line expression systems, animal expression systems are more complex, and potassium currents are affected by other genes in addition to KCNAB3. Additionally, the function of human Kvβ3 might not be fully expressed in the heterologous expression system.
Acknowledgments
Funding: This work was funded by the Key Projects of National Key Research and Development Program of China (No. 2016YFC1306201), National Natural Science Foundation of China for Youth (No. 81701284), Outstanding Young Medical Talents Project of Guangdong Province (No. KJ012019451), Project of Dongguan People’s Hospital (No. K201913), Natural Science Foundation of Guangdong Province (No. 2019A1515011864), Guangzhou Science and Technology Planning Fund (No. 202002030428), and Scientific Research Project of Bureau of Traditional Chinese Medicine of Guangdong Province (No. 20222030).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-436/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-436/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-436/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. KY-D-2021-120-06) granted by the ethics committee of Guangdong Provincial People’s Hospital, in compliance with Guangdong Provincial People’s Hospital guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scheffer IE, Berkovic SF. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain 1997;120:479-90. [Crossref] [PubMed]
- Scheffer IE, Berkovic SF. Generalized (genetic) epilepsy with febrile seizures plus. In: Engel J, Pedley TA, Aicardi J. editors. Epilepsy: a comprehensive textbook. 2nd ed. Philadelphia: Lippincott, Williams and Wilkins, 2008:2553-8.
- Heron SE, Regan BM, Harris RV, et al. Association of SLC32A1 Missense Variants With Genetic Epilepsy With Febrile Seizures Plus. Neurology 2021;96:e2251-60. [Crossref] [PubMed]
- Zhang YH, Burgess R, Malone JP, et al. Genetic epilepsy with febrile seizures plus: Refining the spectrum. Neurology 2017;89:1210-9. [Crossref] [PubMed]
- Singh R, Scheffer IE, Crossland K, et al. Generalized epilepsy with febrile seizures plus: a common childhood-onset genetic epilepsy syndrome. Ann Neurol 1999;45:75-81. [Crossref] [PubMed]
- Singh R, Andermann E, Whitehouse WP, et al. Severe myoclonic epilepsy of infancy: extended spectrum of GEFS+? Epilepsia 2001;42:837-44. [Crossref] [PubMed]
- Scheffer IE, Harkin LA, Dibbens LM, et al. Neonatal epilepsy syndromes and generalized epilepsy with febrile seizures plus (GEFS+). Epilepsia 2005;46:41-7. [Crossref] [PubMed]
- Qu S, Zhou C, Howe R, et al. The K328M substitution in the human GABAA receptor gamma2 subunit causes GEFS+ and premature sudden death in knock-in mice. Neurobiol Dis 2021;152:105296. [Crossref] [PubMed]
- Sfaihi L, Maaloul I, Kmiha S, et al. Febrile seizures: an epidemiological and outcome study of 482 cases. Childs Nerv Syst 2012;28:1779-84. [Crossref] [PubMed]
- Zhang YH. Clinical and molecular genetic study of epilepsy related to fever sensitivity. Chinese Journal of Practical Pediatrics 2015;30:487-91.
- Roemmich AJ, Vu T, Lukacsovich T, et al. Seizure Phenotype and Underlying Cellular Defects in Drosophila Knock-In Models of DS (R1648C) and GEFS+ (R1648H) SCN1A Epilepsy. eNeuro 2021;8:ENEURO.0002-21.2021.
- Ding J, Miao QF, Zhang JW, et al. H258R mutation in KCNAB3 gene in a family with genetic epilepsy and febrile seizures plus. Brain Behav 2020;10:e01859. [Crossref] [PubMed]
- Camfield P, Camfield C. Febrile seizures and genetic epilepsy with febrile seizures plus (GEFS+). Epileptic Disord 2015;17:124-33. [Crossref] [PubMed]
- Zou HF, Zhai QX, Wang LG, et al. Novel KCNQ4 gene mutations contribute to susceptibility of genetic febrile seizures. Int J Clin Exp Pathol 2016;9:12030-40.
- Yu SJ, Bao XH, Yi Y, et al. Clinical analysis of KCNQ3, SCN3A gene variation in a family with generalized epilepsy with febrile seizures plus (report of 1 case). Chinese Journal of Practical Pediatrics 2018;33:924-6.
- Tian Y, Li XJ, Wang XY, et al. Analysis of a family with inherited generalized epilepsy with febrile seizures plus caused by the KCNT2 mutation and literature review. Chinese Journal of Applied Clinical Pediatrics 2021;36:136-9.
- Heinemann SH, Rettig J, Wunder F, et al. Molecular and functional characterization of a rat brain Kv beta 3 potassium channel subunit. FEBS Lett 1995;377:383-9. [Crossref] [PubMed]
- Leicher T, Bähring R, Isbrandt D, et al. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium channel. J Biol Chem 1998;273:35095-101. [Crossref] [PubMed]
- Rettig J, Heinemann SH, Wunder F, et al. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature 1994;369:289-94. [Crossref] [PubMed]
- Bähring R, Vardanyan V, Pongs O. Differential modulation of Kv1 channel-mediated currents by co-expression of Kvbeta3 subunit in a mammalian cell-line. Mol Membr Biol 2004;21:19-25. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)



