
An X-linked PLXNB3 mutation identified in patients with congenital heart disease with neurodevelopmental disabilities
Highlight box
Key findings
• These findings suggest that PLXNB3 and the p.E1440V variant may be related to the pathogenesis of CHD associated with NDD.
What is known and what is new?
• Plexin families are involved in many aspects of heart development and the occurrence of neurodevelopmental anomalies, and gene mutations or abnormalities in the Sema/plexin signaling pathway can cause a variety of cardiovascular diseases.
• In this study, we first identified PLXNB3 as a pathogenic gene in patients with CHD with NDD, our research revealed that p.E1440V variant affected the cardiac muscle contraction and neurodevelopmental pathway, which is involved in the pathogenesis of CHD with NDD.
What is the implication, and what should change now?
• We consider PLXNB3 to be a possible gene marker for diagnosis of CHD with NDD. However, this hypothesis should be verified by more samples from CHD with NDD patients carried PLXNB3 mutations.
Introduction
Congenital heart disease (CHD), one of the most common birth defects, refers to an abnormality of the heart or large blood vessels identified at birth (1). During the period from 1970 to 2017, the global prevalence of CHD gradually increased. The estimated incidence of CHD in Asia is 9.342/1,000 (2), and according to a prospective study, the estimated overall prevalence of CHD in China is 8.98/1,000 (3). CHD is often associated with other neurodevelopmental disabilities (NDD) and genetic or syndromic conditions (4,5): 10% of children with CHD and 50% of those with severe CHD also have NDD (6). Epidemiological studies suggest that environmental or genetic factors are involved in the causation of approximately 20–30% of CHD cases, while other unexplained CHD with NDD cases are believed to have a multifactorial causation (7). In recent years, an increasing number of studies have identified single-gene variants that are closely related to CHD. These single-gene variants mainly pertain to transcription factors, cell signaling and adhesion proteins which are closely related to the development of the heart (8,9).
PLXNB3, a member of the highly conserved plexin family, is the receptor for the axon guiding molecule Sema5A (10). PLXNB3 is a large transmembrane protein; its extracellular domain mainly includes Sema, 3 plexin-semaphorin-integrin (PSI) domains, and 4 plexin, and transcription factor (IPT) repeats, while the intracellular domain is mainly composed of a GTPase-activating protein (GAP) domain and Rho GTPase-binding domain (RBD) insert (11). Plexin families are involved in many aspects of heart development, such as myocardial compaction and trabeculation, neural crest and cardiac outflow tract development, epicardial development, pulmonary vein patterning, autonomic innervation of the heart, and vascular patterning (12). Gene mutations or abnormalities in the Sema/plexin signaling pathway can cause a variety of cardiovascular diseases. For example, in one study, Sema3E/plexinD1 pathway was shown to affect ventricular development by regulating Notch signaling pathway and extracellular cell matrix (ECM) dynamics; mice with abnormal Sema3E/plexinD1 pathway showed left ventricular noncompaction (LVNC) (13). Recent study has reported that the R1229C mutation of PLXND1 is pathogenic in persistent truncus arteriosus (14).
No report yet exists concerning the PLXNB3 mutation in isolated CHD or CHD with NDD. In this study, we performed whole-exome sequencing (WES) on a patient with CHD complicated with neurodevelopmental anomalies and the patient’s family members in order to identify a new pathogenic PLXNB3 mutation which may play an important role in CHD with NDD. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-556/rc).
Methods
Patient recruitment
All children who presented with CHD-associated NDD at the Children’s Hospital of Fudan University (CHFU) were enrolled in the study from May 2015 to December 2020. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Children’s Hospital of Fudan University (code No. 2016121) and written informed consent was obtained from the parents or legal guardians of the children.
WES and Sanger sequencing
Using the QIAamp DNA Blood Mini Kit (Qiagen), genomic DNA for WES and targeted sequencing was extracted from the peripheral blood samples of both parents and individuals. Blood samples were extracted for whole WES by an Illumina Hiseq X Ten platform. The raw data obtained were compared with the reference genome (GRCh37/hg19), and the following 3 parts of information analysis process were completed: sequencing data quality assessment, variant detection, and variant screening and prediction of association with CHD. For the filtered sequencing results, the core family line was used as a whole for genetic analysis, and the 3 aspects of chromosomal dominant inheritance, invisible inheritance, and compound heterozygosity were screened to identify the possible disease-causing variants, with Sanger sequencing being later applied to verify and minimize the false positives. The Sanger sequencing primers were synthesized by Generay Biotech as follows: PLXNB3, forward, 5'- CTCATCCACACCCTGGAGGAGC-3' and reverse, 5'- CTGCGTAGCATGAGCTTGGGG-3'.
De novo homology modeling by AlphaFold and PyRosetta
We firstly used the SWISS-MODEL and CABS-fold servers to model the structure of PLXNB3; however, the obtained models were not acceptable because of the low homology with the template proteins. Subsequently, AlphaFold, the deep learning algorithm developed by DeepMind, was used to predict the model of PLXNB3 in order to obtain more stable wild type (WT) structures. We then performed molecular dynamics (MD) simulations using PyRosetta, the script-based interface for implementing MD simulations algorithm, to obtain the protein structure of the mutant. In addition, Pymol, the molecular graphics system, as used to align and calculate the root-mean-square deviation (RMSD) score between the WT and mutant protein structure.
Cell culture and cell passage
The HEK293T and AC16 cell lines were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Thermo Fisher Scientific) with 10% fetal bovine serum (FBS; Gibco), and 1X penicillin-streptomycin (Gibco) at 37 ℃ in a 5% CO2 atmosphere. The PLXNB3 ORF from the complement DNA (cDNA) library was subcloned into the pcDNA3.1 vector containing the Flag-tag produced the WT PLXNB3 construct. PLXNB3-E1440V was created using the KOD-Plus Mutagenesis Kit (Toyobo). Following the manufacturer’s instructions, Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific) was used to transfect plasmids into cells in Corning culture dishes at a density of 50–70%. Given on the consistent protein expression at 24 and 48 h collected after transfection, the transfected cells were used for further investigation after 24 hours.
Western blotting
The cells were placed on ice after 24-hour transfection. Total proteins were extracted using RIPA buffer and a stop protease inhibitor cocktail (Thermo Fisher Scientific). A bicinchoninic acid (BCA) protein assay kit (Takara Bio) was used to measure the protein concentrations in accordance with the manufacturer’s instructions. Subsequently, polyacrylamide gel was used to isolate an equal number of proteins and transfer them to polyvinylidene difluoride membranes (MilliporeSigma). The membranes were incubated overnight with primary antibodies at 4 ℃ and incubated with secondary antibodies for 2 hours at room temperature after being blocked in 5% skimmed milk for 90 minutes. Antibody against Flag, antibody against GAPDH, and antibody against rabbit were obtained from Cell Signaling Technology.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
HEK293T is a good cell model for exploring mechanisms of gene functions. To investigate the effect of mutations on PLXNB3 function, HEK293T cell RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) and was reverse-transcribed into cDNA using a PrimeScript RT reagent Kit (Takara Bio). cDNA was them amplified by RT-qPCR using the TB Green Premix Ex Taq (Takara Bio). Human plexin-B3, GAPDH, and other signaling pathway–related primers are listed in Table S1.
Scratch wound assay
AC16 cells were plated into 6-well culture dishes. After attainment of 80% confluence, transient transfection was completed with the Flag, WT, and mutant plasmids according to the Lipo3000 (Thermo Fisher Scientific) manufacturer’s protocol. After AC16 cells had attained confluent growth, a wound was produced by using 20-µL pipette tip. Cells were washed twice using phosphate-buffered saline (PBS) to remove floating cells and debris. The wound area was then monitored and measured at 0 and 24 hours.
Flow cytometry for assessment of cell proliferation
Cells were acquired using the Cytofix/CytopermTM Fixation/Permeabilization Solution Kit (BD Biosciences), and then the percentage of Ki67-positive cells was determined. Anti-Ki67 antibody was added (1:500), and the group of tubes without antibody was used as the control. Antibody against Ki67 and antibody against rabbit immunoglobin G (IgG); H+L; Phycoerythrin (PE) were obtained from Abcam.
RNA sequencing
The overexpression efficiency of HEK293T cells transfected with Flag, WT, and mutant plasmids was found to be consistent. Subsequently, transcriptome sequencing and data analysis were performed by Novogene.
Statistical analysis
All data were processed as mean ± standard deviation (SD) values from at least 3 independent experiments. Student t-test and 2-tailed Fisher exact test were used for all analyses. P<0.05 was considered indicative of statistical significance. Figures were created using GraphPad Prism 9.0 (GraphPad Software).
Results
Identification of the PLXNB3 mutation in patients
In a clinic in the Yunnan province of China, we found a proband aged 29 years who had CHD with neuromuscular malformations (Figure 1A). At the age of 6 years, he gradually developed limb weakness, and by the age of 26 years, he was completely unable to walk. In the past 5 years, he had recurrent exacerbations of congestive heart failure and heart enlargement. A 10-year-old child in the family pedigree had similar clinical manifestations. By tracking the medical history, we identified 3 suspected patients in this pedigree who died at the age of 30 years.
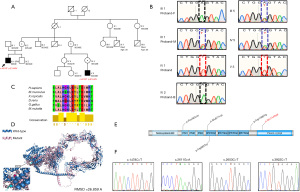
We performed WES to identify the missense mutation in the X-linked PLXNB3 gene (c.A4319T p.E1440V), which was confirmed by Sanger sequencing (Figure 1B, Figure S1). The mutation site was highly conserved among human, mouse, zebrafish, and other species (Figure 1C), which indicated that this missense variant was important for the function of the protein. We used AlphaFold and PyRosetta for de novo homology modeling of PLXNB3 structure to compare the 3-dimensional structure of the WT PLXNB3 protein and the mutant p.E1440V protein. We found that the p.E1440V variant would cause structural change in amino acid, and the RMSD score was 26.859 Å (Figure 1D). The p.E1440V variant was predicted to be possibly pathogenic by Combined Annotation Dependent Depletion (CADD), Polyphen2 algorithm, and Sortig Intolerant From Tolerant (SIFT) program (Table 1).
Table 1
| Gene | Variant type | Variant | Chromosomal position (GRCh37/hg19) | rs ID | CADD (v1.6) | Polyphen2 | SIFT | gnomAD | |
|---|---|---|---|---|---|---|---|---|---|
| Total | East Asian | ||||||||
| PLXNB3 | Missense | c.A4319T, p.E1440V | chrX:153040774 | NA | 28.5 | D | T | Absent | Absent |
NA, not available; CADD, Combined Annotation Dependent Depletion; SIFT, Sortig Intolerant From Tolerant; T, tolerated; gnomAD, The Genome Aggregation Database.
We then recruited 285 patients with sporadic CHD who had concomitant NDD for targeted sequencing. As shown in Table 2, we observed 4 missense mutations which qualified the filtering criteria [Minor Allele Frequency (MAF) <0.01% and CADD >20]. Among these 285 patients, 4 were diagnosed as CHD with NDD (Table 3). In addition, 4 patients were hemizygote boys. The mutation sites were mainly concentrated on the non-Sema domain (Figure 1E). These variants were verified using Sanger sequencing (Figure 1F). To further explore whether these mutations alter the structure of PLXNB3, we used AlphaFold and PyRosetta to model protein structures de novo. All the mutations were found to cause dramatic changes in protein structures (RMSD >10 Å; Figure S2).
Table 2
| Patient ID | Sex | Exon | Chromosomal position (GRCh37/hg19) | Variant | Zygosity | Impact | gnomAD | CADD (v1.6) | |
|---|---|---|---|---|---|---|---|---|---|
| East Asian | Total | ||||||||
| NO_2727 | M | 7 | chrX:153034630 | c.1478C>T, p.P493L | Hemizygote | Missense variant | – | – | 20.5 |
| NO_1097 | M | 16 | chrX:153037343 | c.2611G>A, p.E871K | Hemizygote | Missense variant | – | – | 23.3 |
| NO_1116 | M | 16 | chrX:153037385 | c.2653C>T, p.R885W | Hemizygote | Missense variant | 0 | 6.40E-05 | 25.5 |
| NO_3077 | M | 24 | chrX:153040183 | c.3922C>T, p.R1308W | Hemizygote | Missense variant | – | – | 33 |
gnomAD, The Genome Aggregation Database; CADD, Combined Annotation Dependent Depletion.
Table 3
| Patient ID | Sex | Age (days) | Phenotype | |
|---|---|---|---|---|
| Congenital heart defects | Neurodevelopmental disabilities | |||
| NO_2727 | M | 239 | PDA | Brain atrophy |
| NO_1097 | M | 1666 | TGA | Epilepsy |
| NO_1116 | M | 25 | VSD, ASD | Epilepsy, Hypospadias |
| NO_3077 | M | 186 | TGA, PDA | Brain dysplasia |
CHD, Congenital Heart Defect; PDA, Patent Ductus Arteriosus; TGA, Transposition of Great Arteries; VSD, Ventricular Septal Defect; ASD, Atrial Septal Defect.
The above findings suggested that the X-linked PLXNB3 gene and the p.E1440V variant may play an important role in the occurrence of CHD with NDD.
The influence of the PLXNB3 mutation in migration and proliferation of AC16 cells
In order to observe whether the PLXNB3 mutation affects messenger RNA (mRNA) expression, we performed RT-qPCR by overexpressing the Flag, WT, and mutant plasmids in the AC16 cells. As shown in Figure 2A, the identified possible pathogenic variant was found to have no effect on mRNA expression. We also used western blotting to assess the influence of mutation on protein expression. As shown in Figure 2B, there was no significant difference in protein expression between the WT and the mutant groups.

It was reported that PLXNB3 is linked to cell migration (15). To verify whether the p.E1440V mutant affects cell migration, we used scratch wound assay on AC16 cells. In the AC16 cells transfected with WT plasmid, the migration ability was lower than that of the Flag plasmid, but in the AC16 cells transfected with the p.E1440V mutation plasmid, the migration ability was higher than that of WT plasmid (Figure 2C). To assess the effect of this mutation on cell proliferation, we performed flow cytometry to detect Ki67 expression. As shown in Figure 2D, compared with that in the control group, the percentage of Ki67+ in the WT group was significantly decreased; however, in comparison to that of the WT group, proliferation of AC16 cells of the p.E1440V mutation group was significantly enhanced.
These results showed that this mutation may inhibit cell migration and cell proliferation.
The effect of the PLXNB3 mutation in Notch signaling pathway, cardiac muscle contraction pathway, and neurodevelopmental pathway
We subsequently performed RNA sequencing to detect the changes in the signaling pathway caused by the p.E1440V variant. In comparison to the WT group, there were 1764 differentially expressed genes (|log2 [fold change]|>0 and P value <0.05) in the mutant group, of which 826 were upregulated and 938 were downregulated (Figure 3A). Differentially expressed genes were concentrated in signal pathways such as ribosome, Huntington disease, oxidative phosphorylation, Parkinson disease, Alzheimer disease, nonalcoholic fatty liver disease (NAFLD), and spliceosome and cardiac muscle contraction by KEGG enrichment analysis (Figure 3B). We also found changes in genes related to the Notch signaling pathway during differential gene analysis. Among these, the signaling pathways that were clearly related to heart development included Notch and cardiac muscle contraction pathway. The pathways related to neurodevelopmental diseases included Huntington disease, Parkinson disease, and Alzheimer disease.
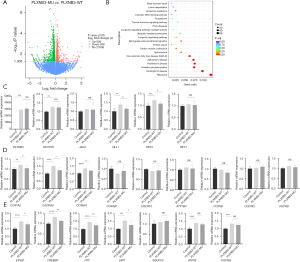
We further verified the effect of the p.E1440V variant on the above pathways by RT-qPCR. We tested some key genes of the Notch pathway (Figure 3C). Compared to PLXNB3 in control group, the PLXNB3 in WT group and mutant groups were overexpressed, and the transfection efficiency of the WT group and mutant group was consistent. In comparison to the control group, the other groups showed significant upregulation of DLL1 and HES1. The mRNA levels of DLL1 and HES1 in the HEK293T cells transfected with the p.E1440V variant plasmid were lower than those of the WT plasmid. We did not find significant differences in NOTCH1, JAG1, and HEY1 between the WT group and the p.E1440V variant group. We next examined some significantly different genes in the cardiac muscle contraction pathway (Figure 3D). We observed a remarkable increase of mRNA expression of only COX7A2, CACNG5 and COX6A1 in the WT group, and significantly lower mRNA expression of only COX7A2, CACNG5 and COX6A1 in the p.E1440V variant group compared to the WT group. To further evaluate whether this mutant influenced the activity of neurodevelopmental pathway, we selected several genes with obvious differences in RT-qPCR (Figure 3E). In the WT group, CREBBP, LRP1, HTT, EP300 showed significant increase relative to the control group, but showed decrease relative to the mutant group. UQCRH, NDUFS3, and ATP5E showed no significant difference between HEK293T cells transfected with the WT plasmid and HEK293T cells transfected with the p.E1440V variant.
The above results indicated that the p.E1440V variant may downregulate the activity of Notch and cardiac muscle contraction pathway. Moreover, it affected the activity of neurodevelopmental-related factors.
Discussion
An accumulating body of evidence has indicated an important role of the plexin family in heart development, neurodevelopment, and cancer progression, but most reports have focused on PLXND1 (12). In this study, we not only identified the PLXNB3 p.E1440V variant in a pedigree with CHD and neurodevelopmental anomalies, but also identified 12 other variants in patients with sporadic CHD. These 13 identified mutations were all located on the non-Sema domain; therefore, we speculate that the non-Sema domain in the protein may be closely related to CHD. Among all the pathogenic mutations identified in patients with sporadic CHD, 4 patients had only CHD, and 7 patients had CHD with NDD. Patients with CHD who have NDD outnumbered those with CHD. Therefore, we speculate that the PLXNB3 mutation may be more dominant in the category of CHD with NDD, which requires further investigations in the CHD population.
In this study, we found that the p.E1440V mutation may play a key role in cardiogenesis and neurogenesis by regulating the Notch signaling pathway, myocardial contraction pathway, and neurodevelopmental pathway. Notch signaling plays an important role in cardiogenesis and disease models. Almost all cell types associated with the cardiovascular system are regulated directly or indirectly by Notch signaling (16). Aberrant Notch signaling always appears to cause disease phenotypes in each system examined (17). Effectors of the Notch signaling pathway (DLL1, HES1) are important for heart development. We found that these genes were reduced in HEK293T cells expressing mutant plasmids. Hes1 is expressed in second heart field (SHF). Moreover, the lack of HES affects SHF and cardiac neural crest cell populations, resulting in defects in the proliferation of the SHF, decreased cardiac neural crest cells, and the inability to fully expand the outflow tract in the early mice embryo (18). In one study, the loss of function of DLL1 in embryos was shown to cause asymmetric development of the heart, with the direction of embryonic turning being random (19). The downregulation of these effectors may lead to the interruption of signaling pathways essential for heart development, leading to heart defects. Hence, we consider PLXNB3 to be a possible gene marker for diagnosis of CHD. However, this hypothesis should be further verified by larger sample of patients with CHD.
The COX7A2 and COX6A1 genes belong to a cytochrome oxidase family, which impairs the systolic and diastolic function of muscle (20). In a previous study, COX7A1 knockout mice showed heart damage with impairment of systolic and diastolic function (21). We found that the mRNA expression levels of COX7A2 and COX6A1 in the mutant group were reduced, which is consistent with the muscle phenotypes of our patients.
Our data suggest a reduced expression level of CREBBP, LRP1, HTT, and EP300 in the mutation group, indicating that the PLXNB3 p.E1440V variant may cause a neurological disease phenotype. LRP1 has been shown to play a key role in the generation of oligodendrocytes, while the lack of LRP1 in the neural stem cells was shown to significantly reduce their ability to differentiate into oligodendrocytes (22). Other research reports that mice with the LRP1 p. C4232R variant had a phenotype of a atrioventricular septal defect with a double-outlet right ventricle, and a similar phenotype can occur with the conditional deletion of LRP1 in cardiac neural crest cells (23). The HTT gene is essential for early neural development. Decreased expression of HTT in neuroepithelial cells was found to increase neuronal cell apoptosis, reduce proliferation, and impair migration ability (24). In patients with Huntington disease caused by a loss of HTT function, the metabolism of cardiomyocytes may be impaired, and neurological symptoms may be accompanied by cardiovascular pathology (25). Previous reports have pointed out that the loss of function of the CREBBP and EP300 genes can lead to Rubinstein-Taybi syndrome (RSTS), which is a congenital disease characterized by intellectual disability, developmental delay, and various malformations (26,27). Deletion or decreased expression of these genes may be related to the occurrence of neurological diseases. Therefore, we speculate that the p.E1440V mutation may be related to the neurodevelopmental defects of the proband.
Normal growth of the embryonic heart is maintained by highly proliferating cardiac tissue cells and a precisely regulated cardiomyocyte proliferation cycle (28). Proliferation and migration of cardiomyocytes are necessary for ventricular trabeculation and cardiac outflow tracts (29). Several studies have shown that PLXNB3 is closely related to cell migration and has a certain effect on cell proliferation. However, the contemporary studies on the effects of PLXNB3 on cell migration and proliferation have been mainly conducted in the context of tumors. PLXNB3 was shown to significantly inhibit glioma cell migration and cell invasion caused by Sema stimulation, but this effect almost disappeared after knockdown of PLXNB3 (30). In the study by Sadanandam et al. (31), knockdown of PLXNB3 in microvascular endothelial cells (HMEC-1) was found to inhibit cell proliferation stimulated by Sema5A. However, there are few reports on the role of PLXNB3 mutations associated with CHD with neuromuscular malformations in cell migration and proliferation. Therefore, we further explored whether the p.E1440V variant affects cell migration and proliferation, and we found that the p.E1440V variant has an inhibitory effect on the migration and proliferation of AC16 cells compared with the WT. HES1 is a transcriptional regulatory factor that plays an important role in cell proliferation, differentiation, and migration. Murata et al. (32) demonstrated that HES1 may promote cell proliferation by altering P27 KIP1 gene expression. Therefore, we speculate that the PLXNB3 p.E1140V variant may affect cell proliferation and migration by reducing the expression of HES1.
Some limitations of our study should be acknowledged. First, the role of changes in cell proliferation and migration induced by PLXNB3 p.E1440V is unclear in CHD with NDD. Second, we didn’t detect the expression changes of PLXNB3 in the patients with CHD with NDD because of difficulty of recalling patients. Therefore, we will try to collect more samples from new CHD with NDD patients carried PLXNB3 mutations to detect the PLXNB3 expression and explore the pathogenic role of PLXNB3 mutations in isolated CHD or CHD with NDD.
Conclusions
In this study, we identified a new pathogenic PLXNB3 p.E1440V mutation in a family with CHD and NDD which may expand the spectrum of PLXNB3 variants associated with CHD with NDD. Our study shows that the functional changes caused by PLXNB3 pathogenic mutation may be a potential genetic mechanism of CHD with NDD.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (Nos. 2021YFC2701000 and 2016YFC1000500), the National Natural Science Foundation of China (Nos. 81873482, 81873483, and 81800282), the Shanghai Basic Research Project of Science and Technology Innovation Action Plan (No. 20JC1418300), and the CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-5-002).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-556/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-556/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-556/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Children’s Hospital of Fudan University (code No. 2016121) and written informed consent was obtained from the parents or legal guardians of the children.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van der Linde D, Konings EE, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol 2011;58:2241-7. [Crossref] [PubMed]
- Liu Y, Chen S, Zühlke L, et al. Global birth prevalence of congenital heart defects 1970-2017: updated systematic review and meta-analysis of 260 studies. Int J Epidemiol 2019;48:455-63. [Crossref] [PubMed]
- Zhao QM, Liu F, Wu L, et al. Prevalence of Congenital Heart Disease at Live Birth in China. J Pediatr 2019;204:53-8. [Crossref] [PubMed]
- Dowden L, Tucker D, Morgan S, et al. Contribution of Congenital Heart Disorders Associated With Copy Number Variants in Mediating Risk for Brain Developmental Disorders: Evidence From 20-Year Retrospective Cohort Study. Front Cardiovasc Med 2021;8:655463. [Crossref] [PubMed]
- Owen M, Shevell M, Majnemer A, et al. Abnormal brain structure and function in newborns with complex congenital heart defects before open heart surgery: a review of the evidence. J Child Neurol 2011;26:743-55. [Crossref] [PubMed]
- Marino BS, Lipkin PH, Newburger JW, et al. Neurodevelopmental outcomes in children with congenital heart disease: evaluation and management: a scientific statement from the American Heart Association. Circulation 2012;126:1143-72. [Crossref] [PubMed]
- Robinson J, Uzun O, Loh NR, et al. The association of neurodevelopmental abnormalities, congenital heart and renal defects in a tuberous sclerosis complex patient cohort. BMC Med 2022;20:123. [Crossref] [PubMed]
- Kodo K, Uchida K, Yamagishi H. Genetic and Cellular Interaction During Cardiovascular Development Implicated in Congenital Heart Diseases. Front Cardiovasc Med 2021;8:653244. [Crossref] [PubMed]
- Davis K, Azarcon P, Hickenlooper S, et al. The role of demethylases in cardiac development and disease. J Mol Cell Cardiol 2021;158:89-100. [Crossref] [PubMed]
- Artigiani S, Conrotto P, Fazzari P, et al. Plexin-B3 is a functional receptor for semaphorin 5A. EMBO Rep 2004;5:710-4. [Crossref] [PubMed]
- Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol 2005;6:789-800. [Crossref] [PubMed]
- Zhang YF, Zhang Y, Jia DD, et al. Insights into the regulatory role of Plexin D1 signalling in cardiovascular development and diseases. J Cell Mol Med 2021;25:4183-94. [Crossref] [PubMed]
- Sandireddy R, Cibi DM, Gupta P, et al. Semaphorin 3E/PlexinD1 signaling is required for cardiac ventricular compaction. JCI Insight 2019;4:125908. [Crossref] [PubMed]
- Ta-Shma A, Pierri CL, Stepensky P, et al. Isolated truncus arteriosus associated with a mutation in the plexin-D1 gene. Am J Med Genet A 2013;161A:3115-20. [Crossref] [PubMed]
- Kuhlmann L, Govindarajan M, Mejia-Guerrero S, et al. Glycoproteomics Identifies Plexin-B3 as a Targetable Cell Surface Protein Required for the Growth and Invasion of Triple-Negative Breast Cancer Cells. J Proteome Res 2022;21:2224-36. [Crossref] [PubMed]
- Gomez AH, Joshi S, Yang Y, et al. Bioengineering Systems for Modulating Notch Signaling in Cardiovascular Development, Disease, and Regeneration. J Cardiovasc Dev Dis 2021;8:125. [Crossref] [PubMed]
- Salazar JL, Yang SA, Yamamoto S. Post-Developmental Roles of Notch Signaling in the Nervous System. Biomolecules 2020;10:985. [Crossref] [PubMed]
- Rochais F, Dandonneau M, Mesbah K, et al. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS One 2009;4:e6267. [Crossref] [PubMed]
- Przemeck GK, Heinzmann U, Beckers J, et al. Node and midline defects are associated with left-right development in Delta1 mutant embryos. Development 2003;130:3-13. [Crossref] [PubMed]
- Lenka N, Vijayasarathy C, Mullick J, et al. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol 1998;61:309-44. [Crossref] [PubMed]
- Hüttemann M, Klewer S, Lee I, et al. Mice deleted for heart-type cytochrome c oxidase subunit 7a1 develop dilated cardiomyopathy. Mitochondrion 2012;12:294-304. [Crossref] [PubMed]
- Schäfer I, Kaisler J, Scheller A, et al. Conditional Deletion of LRP1 Leads to Progressive Loss of Recombined NG2-Expressing Oligodendrocyte Precursor Cells in a Novel Mouse Model. Cells 2019;8:1550. [Crossref] [PubMed]
- Lin JI, Feinstein TN, Jha A, et al. Mutation of LRP1 in cardiac neural crest cells causes congenital heart defects by perturbing outflow lengthening. Commun Biol 2020;3:312. [Crossref] [PubMed]
- Tong Y, Ha TJ, Liu L, et al. Spatial and temporal requirements for huntingtin (Htt) in neuronal migration and survival during brain development. J Neurosci 2011;31:14794-9. [Crossref] [PubMed]
- Tomczyk M, Glaser T, Ulrich H, et al. Huntingtin protein maintains balanced energetics in mouse cardiomyocytes. Nucleosides Nucleotides Nucleic Acids 2022;41:231-8. [Crossref] [PubMed]
- Roelfsema JH, White SJ, Ariyürek Y, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet 2005;76:572-80. [Crossref] [PubMed]
- Wang Q, Xu W, Liu Y, et al. A Novel CREBBP in-Frame Deletion Variant in a Chinese Girl with Atypical Rubinstein-Taybi Syndrome Phenotypes. J Mol Neurosci 2021;71:607-12. [Crossref] [PubMed]
- Günthel M, Barnett P, Christoffels VM. Development, Proliferation, and Growth of the Mammalian Heart. Mol Ther 2018;26:1599-609. [Crossref] [PubMed]
- Sanchez-Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Semin Cell Dev Biol 2010;21:929-35. [Crossref] [PubMed]
- Li X, Lee AY. Semaphorin 5A and plexin-B3 inhibit human glioma cell motility through RhoGDIalpha-mediated inactivation of Rac1 GTPase. J Biol Chem 2010;285:32436-45. [Crossref] [PubMed]
- Sadanandam A, Rosenbaugh EG, Singh S, et al. Semaphorin 5A promotes angiogenesis by increasing endothelial cell proliferation, migration, and decreasing apoptosis. Microvasc Res 2010;79:1-9. [Crossref] [PubMed]
- Murata K, Hattori M, Hirai N, et al. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol 2005;25:4262-71. [Crossref] [PubMed]


