
Genome-wide association study identifies WWC2 as a possible locus associated with persistent pulmonary hypertension of the newborn in the Thai population
Highlight box
Key findings
• A genome-wide association study (GWAS) showed that a single nucleotide polymorphism (SNP) rs149768622 T>C in the WWC2 gene was associated with persistent pulmonary hypertension of the newborn (PPHN) (P=3.76E-08, odds ratio 13.24, and 95% CI: 3.91–44.78). WWC2 protein plays a role in the formation of critical alveoli.
What is known and what is new?
• PPHN is attributable to genetic polymorphisms of multiple genes and environment. SNPs in genes, such as CRHR1, CPS1, EDN1, NOTCH3, EPAS1, and SMAD9 were reportedly associated with PPHN.
• This study is the first to utilize GWAS in searching for genetic polymorphisms associated with PPHN.
• WWC gene is a possible candidate gene modulating the risk for PPHN.
What is the implication?
• Further studies regarding functional effects of WWC2 gene in PPHN should be carried out.
• Genetic testing of identified genetic variants in at risk families provides information whether the next children should be closed monitoring during perinatal period.
Introduction
Persistent pulmonary hypertension of the newborn (PPHN) is a serious respiratory disease characterized by an elevation in pulmonary vascular resistance in the neonatal pulmonary circulation (1). PPHN results from a failure of pulmonary vasculature relaxation, which in turn results in shunting of non-oxygenated blood from the pulmonary to the systemic circulation at birth. Recent studies have reported incidences of PPHN ranging between 1.2 and 4.6 per 1,000 live-births (2,3). There are various etiologies of PPHN, most commonly meconium aspiration syndrome, transient tachypnea of the newborn, and respiratory distress syndrome. PPHN also manifests in patients with congenital anomalies (e.g., congenital diaphragmatic hernia or lung hypoplasia) (2,4,5). Other causes of PPHN that are unrelated to respiratory conditions include sepsis, birth asphyxia, and maternal drug/smoke exposure (6,7). A small fraction of patients without any of the above-mentioned risk factors are diagnosed with idiopathic PPHN.
PPHN is a complex disease that is attributable to genetic variants of multiple genes and environmental influences. Proper lung development depends on timely action from various genes, and mutations in any of these genes can result in maldevelopment of abnormal growth in the lungs, which can lead to pulmonary hypertension (8). Genetic mutations also can adversely result in either maladaptation or abnormal growth of the lung, which ultimately leading to pulmonary hypertension (8). To date, little is conclusively known concerning genetic involvement with PPHN. Numerous studies have examined the genes that encode certain proteins related to proper lung development, such as vasodilators, vasoconstrictors, surfactants, vascular growth factors, and serotonin receptors, from which studies only a few genetic loci have been identified in patients with PPHN, including the CRHR1, CPS1, EDN1, NOTCH3, EPAS1, TTLL3, ITGAM, TBX4, and SMAD9 genes (9-16). Few of these studies used a genome-wide approaches (13,14). Genetic studies in individuals with pulmonary arterial hypertension (PAH) are also helpful in understanding the contributions to PPHN, although the disease mechanisms of this condition are slightly different from PAH (17,18). An extensive literature review by Morrell et al. summarized the presence of causal mutations of genes, identifying BMPR2, EIL2AK4, TBX4, ATP13A3, GDF2, SOX17, AQP1, ACVRL1, SMAD9, ENG, KCNK3, CAV1, SMAD4, SMAD1, KLF2, BMPR1B, and KCNA51 in adult or childhood onset PAH (19).
A genome-wide association study (GWAS) is a study in which genotypes of multiple genetic loci of the human genome of patients and healthy controls are analyzed to search for specific genetic variations that are associated with a particular disease in order to gain a better understand of how these genes contribute to the disease. In this study, we performed a GWAS to search for genetic variants associated with PPHN. We also summarized the genetic findings related to PPHN from previous studies. We present the following article in accordance with the STREGA reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-280/rc).
Methods
Study population
The GWAS data were obtained from a previous study in which we investigated associations between selected genes involved in the pathogenesis of PPHN (16). A total of 54 infants, who were diagnosed with PPHN between January 2013 and December 2019, were recruited into this case-control study. The inclusion criteria were infants who were diagnosed with PPHN and were born to mothers with gestational age of ≥35 weeks and birth weight of ≥2,000 g. Infants who had congenital malformations or recognizable genetic diseases, e.g., congenital cyanotic heart disease, congenital diaphragmatic hernia, or chromosomal disorders, were excluded from the study. The diagnostic criteria for PPHN were refractory hypoxia and one of the three following conditions: (I) documented pulmonary hypertension as defined by echocardiographic evidence of elevated pulmonary pressure (right to left or bidirectional shunt; (II) a pre- to post ductal partial pressure of oxygen gradients equal to or greater than 20 mmHg; and/or (III) a pulse oximetry oxygen saturation gradient equal to or greater than 10% (20). For a control group, we recruited 100 healthy newborns. In addition, we included 233 healthy adults who had participated in the Thai SNP Database Project of Ramathibodi Hospital, 333 control participants were finally included in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of Hat Yai Hospital (No. 24/2562) and informed consent was taken from the patients’ parents. A flow diagram of the analysis is shown in Figure 1.
Genotyping and quality control in the GWAS
We performed single nucleotide polymorphism (SNP) genotyping by using an Illumina Asian Screening Array-24 v1.0 BeadChip, following the Illumina Infinium HTS Assay protocol (Illumina, San Diego, CA, USA). The array platform contained 659,184 SNPs probes across the genome. We applied standard SNP quality control filters to exclude SNPs with a low call rate of <98%, a missing genotype rate per individual of >10%, a missing genotype rate per SNPs of >5%, gender inconsistency based on the heterozygosity rate of the X chromosome (maleTh =0.8, femaleTh =0.2), Hardy-Weinberg equilibrium test (P<1E-06), minor allele frequency (MAF) of each SNP of <0.01, and genome-wide estimates of identity-by-descent (pihat) ≥0.185 (3rd generation). We were left with 443,063 variants after quality control.
Statistical analysis
This study used the FaST-LMM (V2.02.20121124) and R Project for general statistical analysis. A Manhattan plot of −log10P was generated using the R package. Quantile-quantile plots (Q-Q plots) were used to assess the overall quality of the GWAS. Significant SNPs with a strong association for PPHN were defined as having statistical significance at the genome-wide association P value threshold of <1E-08 and SNPs with suggestive evidence for PPHN with a P value threshold of <1E-05.
Results
Among the 54 infants with PPHN, approximately two-thirds had meconium aspiration syndrome or transient tachypnea of the newborn, with frequencies of 46.3% and 24.1%, respectively. Idiopathic PPHN was diagnosed in one patient (1.8%). The mean gestational age was 39.5±2.1 weeks, the mean birthweight was 3,105±471 g, and the median Apgar scores at 1 minute and 5 minutes were 8 and 9, respectively. A summary of the baseline characteristics is shown in Table 1.
Table 1
| Characteristic | Values |
|---|---|
| Gestational age (weeks) | 39.5±2.1 |
| Birth weight (g) | 3,105±471 |
| Age | |
| PPHN subjects from prospective recruitment (days) (n=25) | 2 (1.0–4.0) |
| PPHN subjects from retrospective data (years) (n=20) | 2.3 (1.0–3.5) |
| Gender | |
| Male | 32 (59.3) |
| Female | 22 (40.7) |
| Perinatal history | |
| Apgar score at 1 minute | 8 (7–9) |
| Apgar score at 5 minutes | 9 (8–9) |
| Causes of PPHN | |
| Meconium aspiration syndrome | 25 (46.3) |
| Transient tachypnea of the newborn | 13 (24.1) |
| Congenital pneumonia | 7 (13.0) |
| Respiratory distress syndrome | 5 (9.3) |
| Neonatal sepsis | 2 (3.7) |
| Idiopathic PPHN | 1 (1.9) |
| Birth asphyxia | 1 (1.9) |
| Treatment modalities of PPHN | |
| High-frequency oscillatory ventilation | 38 (70.4) |
| Intravenous iloprost | 15 (27.8) |
| Intragastric sildenafil | 39 (72.2) |
| Inhaled nitric oxide | 46 (85.2) |
| Intravenous milrinone | 25 (46.3) |
| Hydrocortisone | 23 (42.6) |
| Sedation | 54 (100.0) |
| Analgesia | 54 (100.0) |
| Inotropic agents | 54 (100.0) |
| Outcomes | |
| Length of supplemental oxygenation (days) | 24 (16–30) |
| Length of hospital stay (days) | 22 (16–34) |
| In-hospital mortality | 2 (3.7) |
Data are presented as mean ± standard deviation, median (interquartile range), or n (%). PPHN, persistent pulmonary hypertension of the newborn.
SNP association analysis
The GWAS results for the 443,063 markers that were included in the further analysis of this study showed good overall quality of the GWAS. The genomic inflation factor (λ) was 1.016 (Figure 2A). In this study, the rs149768622 T>C SNP, which is located in the intronic area of the WW and C2 Domain Containing 2 (WWC2) gene on chromosome 4 (4q35.1) had the highest P value (P=3.76E-08) and odds ratio (OR) 13.24 [95% confidence interval (CI): 3.91–44.78] (Figure 2B). The variants at the rs78232913 (LOC102723906/LOC105377599), rs147741304 (intergenic area), rs112876885 (CADM4), rs56956847 (intergenic area), rs191330351 (GPM6A), rs56074206 (CIT), rs7134357 (RIMBP2), rs145547469 (LOC105374510), rs10239513 (LOC105375193), rs11764121 (PTPRN2), rs144344757 (CDK14), rs187729743 (LCORL) and rs79741635 (intergenic area) loci showed suggestive evidence of associations with PPHN (P value <1E-05). The significant findings of our GWAS are summarized in Table 2.
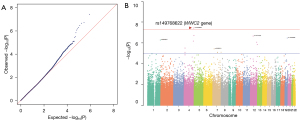
Table 2
| Chr | SNP | Gene (protein) | Protein function | Ref. allele | Risk allele | MAF | Functional consequence | GWAS | ||
|---|---|---|---|---|---|---|---|---|---|---|
| PPHN | Control | OR | P | |||||||
| 4q35.1 | rs149768622 | WWC2 (WW and C2 Domain Containing 2) | Modulator in Hippo pathway signaling | T | C | 0.074 | 0.006 | Intron variant | 13.24 (3.91–44.78) | 3.76E-08 |
| 4q35.2 | rs78232913 | LOC102723906; LOC105377599 | Uncharacterized gene thought to produce ncRNA | G | A | 0.065 | 0.005 | LOC102723906: intron variant; LOC105377599: 2KB upstream variant | 15.32 (3.89–60.19) | 1.93E-07 |
| 12q13.3 | rs147741304 | None* | None | G | A | 0.056 | 0.003 | NA | 19.47 (3.88–97.97) | 2.19E-07 |
| 19q13.31 | rs112876885 | CADM4 (Cell Adhesion Molecule 4); LOC105372411 | Cell-cell adhesion | A | G | 0.056 | 0.003 | CADM4: Missense variant; LOC105372411: intron variant | 19.53 (3.89–98.08) | 3.39E-07 |
| 1p21.2 | rs56956847 | None** | None | A | G | 0.129 | 0.027 | NA | 5.36 (2.58–11.14) | 5.24E-07 |
| 4q34.2 | rs191330351 | GPM6A (Glycoprotein M6A) | Calcium channel activity, neuronal migration, stem cell differentiation | T | C | 0.129 | 0.026 | Intron variant | 5.69 (2.71–11.94 | 6.67E-07 |
| 12q24.23 | rs56074206 | CIT (Citron Rho-Interacting Serine/Threonine Kinase); MIR1178 (MicroRNA 1178) | Cell division, cytokinesis, development of central nervous system | G | A | 0.129 | 0.029 | CIT: intron variant; MIR1178: 2KB upstream variant | 5.06 (2.45–10.42) | 8.52E-07 |
| 12q24.33 | rs7134357 | RIMBP2 (RIMS binding protein 2) | Synaptic transmission | G | A | 0.194 | 0.059 | Intron variant | 3.87 (2.18–6.88) | 1.43E-06 |
| 4p15.31 | rs145547469 | LOC105374510 | Uncharacterized gene thought to produce ncRNA | T | C | 0.111 | 0.023 | Intron variant | 5.38 (2.44–11.83) | 3.11E-06 |
| 7p15.3 | rs10239513 | LOC105375193 | Uncharacterized gene thought to produce ncRNA | T | C | 0.306 | 0.132 | 2KB upstream variant | 2.93 (1.84–5.67) | 3.74E-06 |
| 7q36.3 | rs11764121 | PTPRN2 (Protein Tyrosine Phosphatase Receptor Type N2) | Involved in vesicle-mediated secretory process. May be involved in the regulation of insulin secretion | A | G | 0.083 | 0.015 | Intron variant | 5.96 (2.37–15.04) | 7.76E-06 |
| 7q21.13 | rs144344757 | CDK14 (Cyclin Dependent Kinase 14) | Regulator of cell cycle progression and cell proliferation, cell-cycle Regulator of Wnt signaling pathway | A | G | 0.129 | 0.032 | Intron variant | 4.57 (2.25–9.05) | 7.79E-06 |
| 4p15.31 | rs187729743 | LCORL (Ligand Dependent Nuclear Receptor Corepressor Like) | May act as transcription activator | C | T | 0.065 | 0.008 | Intron variant | 9.16 (2.85–29.42) | 7.87E-06 |
| 7q21.13 | rs79741635 | None*** | None | A | G | 0.129 | 0.032 | NA | 4.57 (2.49–9.31) | 8.40E-06 |
*, an intergenic region lies between the KRT82 and KRT75 genes; **, an intergenic region lies between the GRM2 and RGS7 genes; ***, an intergenic region lies between the CLDN12 and CKD14 genes. Chr, chromosome; SNP, single nucleotide polymorphism; MAF, minor allele frequency; PPHN, persistent pulmonary hypertension of the newborn; GWAS, genome-wide association study; OR, odds ratio; P, P value of heterogeneity; T, thymine; C, cytosine; ncRNA, noncoding ribonucleic acid; G, guanine; A, adenine; NA, not available.
For genotype distribution and allele frequency, the allele frequency of the minor allele “C” of rs149768622 in the WWC2 gene showed a significant difference between the PPHN and control groups (0.074 vs. 0.006, respectively). In addition to WWC2, the suggestive candidate polymorphisms of other genes are summarized in Table 2.
Gene network analysis of candidate genes from the GWAS
A gene network of 8 candidate genes in this study (1 strong evidence, the WWC2 gene, and 7 with suggestive evidence, the CADM4, GPM6A, CIT, RIMBP2, PTPRN2, CDK14, and LCORL genes) and previous genes that were reported to be related to PPHN were analyzed. Genes within the biological network of the WWC2 gene were also included (Figure S1).
Discussion
A number of genetic studies have been conducted to identify genetic factors contributing to PPHN. Most of these studies have focused on evaluating selective SNPs on genes associated with the biological pathways related to the pathogenic mechanisms that contribute to PPHN, such as pulmonary surfactants, endothelin, phosphodiesterase, enzymes in the corticosteroid pathway, and enzymes in the urea cycle pathway. Genetic associations have been identified on multiple genomic loci across the genome. As PPHN and PAH result from the contributions of multiple genetic variants in conjunction with environmental influences, they are considered a complex disease. For a complex disease, GWAS is a cost-effective method for studying multiple genetic variants across the genomes of large numbers of individuals to identify associations between genotypes and a phenotype. GWAS is typically carried out by genotyping of several hundred thousand SNPs using high throughput technologies (e.g., microarray, next generation sequencing). Unlike traditional gene mapping methods, GWAS can be performed on collections of individuals with unknown ancestry. Our study is the first to utilize GWAS to search for genetic variations associated with PPHN. As most of the GWAS identified SNPs that were present in non-coding regions, a functional SNP would be the most likely to affect gene regulation by altering the binding of some transcription factors. In this pilot study, we also identified rs149768622 in the intron 1 of the WWC2 gene as a strong candidate for association with PPHN at the genome-wide significance level (P<1E-08) and 13 SNPs from 10 genes (LOC102723906/LOC105377599, CADM4, GPM6A, CIT, RIMBP2, LOC105374510, LOC105375193, PTPRN2, CDK14, and LCORL) and 2 intergenic areas as having suggestive evidence for associations with PPHN.
The WWC2 gene encodes a member of the WW-and-C2-domain-containing family of proteins. The WWC2 gene is expressed in various human tissues, including the lungs, thyroid, uterus, and liver. Its expression is high in lung tissue (21). The WWC2 protein is an upstream component of the Hippo signaling pathway, which controls organ size in animals through the regulation of cell regulation and apoptosis. In mice, the WWC2 gene plays role in controlling cell differentiation, cell proliferation, and organ growth. In human beings, experimental data have indicated that the protein plays a role in the formation of critical alveoli, which usually occurs postnatally (22). The formation of critical alveoli consists of 2 steps, alveolar septation and alveologenesis. The first step subdivides saccules into alveoli lined by flat type 1 (AT1) cells (i.e., pneumocyte type I) and surfactant-secreting type 2 (AT2) cells (i.e., pneumocyte type II). Pneumocyte type I cells contribute about 95% of the alveolar surface where passive gas exchange between alveoli and capillaries takes place. As a main function of pneumocyte type I cells is for gas exchange, damage or malfunction of these cells could result in the development of neonatal hypoxia and subsequent PPHN. Our study found that rs149768622 T>C, which is located in the first intron of the WWC2 gene, was present in significantly higher amounts in patients with PPHN than in the healthy controls. YAP (Yes1 Associated Transcriptional Regulator) is a protein involved in the Hippo signaling pathway that has been experimentally determined to be co-expressed with the WWC2 protein (Figure S1). Bertero et al. (23) found that YAP/TAZ-dependent activation of glutaminolysis and anaplerosis is a pathogenic mechanism by which vessel stiffness can stimulate cellular proliferation in pulmonary hypertension. YAP/TAZ forms a complex with TEAD, a transcriptional enhancer factor, which also plays a key role in the Hippo signaling pathway. Duong et al. (22) suggested that TEAD is enriched in AT1 cells and directs AT1 cell maturation. Their experiment found that the WWC2 gene had the highest TEAD motif enrichment in AT1 cells. The WWC2 gene is likely involved in physiologic changes at birth from air-inflated stretching and AT1 cell differentiation via interactions with the Hippo pathway. The WWC2 gene is likely to play an essential role in alveolar development after birth, it is possible that WWC2 is a candidate gene for the pathogenesis of PPHN.
Our analysis also found 7 suggestive risk genes, namely the CADM4, GPM6A, CIT, RIMBP2, CDK14, PTPRN2, and LCORL genes. The functions of the proteins encoded from these genes are summarized in Table 2. Interestingly, one of these genes was the PTPRN2 gene, which encodes a member of the protein tyrosine phosphatase (PTP) family. The PTPs remove phosphate groups from tyrosine residues, thereby influencing signal transduction and cellular processes such as cell metabolism, motility and survival. Chen et al. (24) studied epigenome-wide associations between smoke/allergen exposure and the development of asthma and chronic obstructive pulmonary disease, and found that hypomethylated PTPRN2 was associated with rapid lung function.
Recently, a number of genes involved in the pathogenesis of PPHN have been identified. The majority of these studies have focused on selected genes. A summary of previous studies regarding the genes involved in PPHN is shown in Table 3. The gene most reported to be involved in PPHN is the CPS1 gene, which encodes the carbamoyl phosphate synthase enzyme in the urea cycle. Pearson et al. (9) reported a missense variant, T1405N, (i.e., rs7422339 or rs1047891) in a patient with PPHN and a lower plasma concentration of arginine, which is an intermediate in the urea cycle. In the urea cycle, nitric oxide is synthesized during the conversion of arginine to citrulline. The study suggested that low concentrations of intermediates in the urea cycle and nitric oxide precursors may be associated with PPHN. Kaluarachchi et al. (12) also reported 3 SNPs, rs41272673, rs4399666, and rs2287599, in the CPS1 gene, in US patients with PPHN with low tyrosine levels but high phenylalanine levels. El-Khazragy et al. (15) found that the rs4399666 variant in the CPS1 gene was also significantly associated with PPHN in Egyptian infants. Byers et al. (10) studied 32 SNPs in 12 candidate genes involving the pathophysiology of PPHN and found that rs4458044 in the corticotropin-releasing hormone receptor 1 (CRHR1) gene was significantly associated with PPHN. They found that the level of 17-hydroxyprogesterone in infants with PPHN was significantly higher than in the controls. Approximately 14% of the infants in the PPHN group were also found to have congenital adrenal hyperplasia. Mei et al. (11) observed significantly higher GT and TT genotypes in the rs2070699 variant in the endothelin 1 (EDN1) gene and higher plasma levels of endothelin-1 in patients with PPHN. Liu et al. (13) investigated genetic variants associated with 25 reported PPHN/PAH-related genes by panel exome sequencing in 74 PPHN infants. They found that 3 SNPs (rs192759073, rs1047883 and rs2229589) in CPS1 and one SNP (rs1044008) in NOTCH3 were significantly associated with PPHN (P<0.05). Wang et al. (14) found that the p.E317K and p.P777S variants in the tubulin tyrosine ligase-like family member 3 (TTLL3) gene and the p.E1071D variant in the integrin subunit alpha M (ITGAM) gene were associated with PPHN in high altitude areas (Tibet and Han in China). Recently, we found that the rs17034984 variant in the EPAS1 gene, which encodes either the endothelial PAS domain-containing protein 1 or the HIF-2α protein was a possible genetic susceptibility locus associated with PPHN (16). In this GWAS analysis, we found that rs17034984 had nearly reached statistical significance at the genome-wide association P value threshold of <1E-08 (P=1.97E-05), it failed to reach the value, probably due to the dilution effect from the SNPs being tested over the whole genome.
Table 3
| Reference (years) | No. of PPHN cases | Study methodology | Gene (Chr) | Protein | Variant | ||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Protein function | Role in PPHN/ PAH | Name | Functional consequence | |||||
| Pearson et al. 2001 (9) | 31 | Study of 3 polymorphisms of the CPS1 gene using single-strand conformation polymorphism analysis | CPS1 (2q34) | Carbamoyl-phosphate synthetase 1 | Enzyme in the first step of the urea cycle | Nitric oxide is endogenously synthesized from a precursor L-arginine by nitric oxide synthase. L-arginine is an amino acid supplied by urea cycle | rs7422339 (rs1047891) | Missense variant | |
| Byers et al. 2012 (10) | 88 | Family-based candidate gene study (32 SNPs in 12 genes (BMPR2, SFTPB, EPAS1, PDE5A, SOD3, NR3C1, CRHBP, VEGFA, NOS, SFTPC, SFTPD, CRHR1) using TaqMan-based quantitative PCR | CRHR1 (17q21.31) | Corticotropin Releasing Hormone Receptor 1 | A G-protein coupled receptor for corticotropin-releasing factor and urocortin. Required for normal embryonic development of the adrenal gland and for normal hormonal responses to stress | Downstream of rs4458044 is a transcription factor binding site for PPAR-γ. PPAR-γ is a regulator of pulmonary smooth muscle cell proliferation and vascular tone. Decreased expression of PPAR-γ was observed in patients with pulmonary hypertension and neonatal chronic lung disease. Fetal steroid production is also essential for in utero lung development and ex utero pulmonary transition | rs4458044 | Intron variant | |
| Mei et al. 2016 (11) | 55 | Candidate gene sequencing (EDN1, ACE, VEGFA, NOS3 genes) using next generation sequencing | EDN1 (6p24.1) | Endothelin 1 | A potent vasoconstrictor and mitogen for smooth muscles | Endothelin 1 promotes vasoconstriction, which affects pulmonary vasculature (25) | rs2070699 | Intron variant | |
| Kaluarachchi et al. 2018 (12) | 77 | Family-based candidate gene study (48 SNPs in 6 urea cycle genes: CPS1, NAGS, ASS, ASL, ARG1, OTC) using TaqMan-based quantitative PCR | CPS1 (2q34) | Same as above | Same as above | Same as above | rs41272673 | Intron variant | |
| rs4399666 | Intron variant | ||||||||
| rs2287599 | Synonymous variant | ||||||||
| Liu et al. 2019 (13) | 115 | Clinical exome sequencing panel (n=115) then SNP analysis for disease-causing variants in 25 PPHN/PAH genes (n=74) | CPS1 (2q34) | Same as above | Same as above | Same as above | rs192759073 | Synonymous variant | |
| rs1047883 | Missense variant | ||||||||
| rs2229589 | Synonymous variant | ||||||||
| NOTCH3 (19p13.12) | Notch receptor 3 | A receptor that belongs to the Notch signaling pathway, which plays roles in the regulation of cell proliferation, differentiation, differentiation and apoptosis cell-fate determination | Expression of NOTCH3 was increased in the pulmonary artery smooth muscle cells of PAH patients. Heterozygous variants might affect cell proliferation and NOTCH3-HES5 signaling resulting in PAH (26) | rs1044008 | Synonymous variant | ||||
| Wang et al. 2020 (14) | 100 | Whole exome sequencing (n=20) then gene panel sequencing of 237 genes (n=80) | TTLL3 (3p25.3) | Tubulin tyrosine ligase-like family member 3 | Protein with ligase activity that catalyzes ATP-dependent restoration of tyrosine to the C-terminus of α-tubulin. Tubulin is polymerized into microtubules | Abnormality in the detyrosination/tyrosination cycle of tubulin is involved in the development of hypertension. Reduced adhesion of induced pluripotent stem cell-derived endothelial cells obtained from a patient with familial PAH was observed | p.E317K | Missense variant | |
| p.P777S | Missense variant | ||||||||
| ITGAM (16p11.2) | Integrin subunit alpha M | Integral membrane protein involved in adhesive interactions of monocytes, macrophages, and granulocytes | Unclear | p.E1071D | Missense variant | ||||
| El-Khazragy et al. 2021 (15) | 150 | Specific SNP analysis (rs4399666) using TaqMan-based quantitative PCR | CPS1 (2q34) | Same as above | Same as above | Same as above | rs4399666 | Intron variant | |
| Nakwan et al. 2022 (16) | 45 | SNP microarray | EPAS1 (2p21) | Endothelial PAS Domain Protein1 or HIF-2α protein | A transcription factor involved in the induction of oxygen regulated genes. It regulates erythropoiesis and physiological responses to hypoxia through the HIFs signaling pathway | HIF-2α regulates the vascular endothelial growth factor expression, which may be implicated in the development of blood vessels and the tubular system of the lung. HIF-2α stability is involved in arginase expression and nitric oxide homeostasis. Endothelial HIF-2α could induce pulmonary hypertension through an increased expression of endothelin-1 and a decrease in vasodilatory apelin receptor signaling | rs17034984 | Intron variant | |
PPHN, persistent pulmonary hypertension of the newborn; Chr, chromosome; PAH, pulmonary arterial hypertension; PPAR-γ, peroxisome proliferator-activated receptor γ; SNP, single nucleotide polymorphism; PCR, polymerase chain reaction; ATP, adenosine triphosphate; HIF-2α, hypoxia-inducible factor 2 alpha.
GWAS is a high-throughput genotyping test that evaluates a large set of SNPs across the human genome. In this study, we used a case-control study design to compare the frequency of SNPs between patients with PPHN and a matched control group without PPHN. We found that significant genetic variants were observed more frequently in patients with PPHN than in the controls. These variants may either be causative or more likely associated (in ‘linkage disequilibrium apostrophe’) with the causative mutation. There are various implications these GWAS results could have in terms of their clinical applications for PPHN. One possible clinical application of these genetic associations would be to provide predictive and prognostic information in the presence of other risk factors for PPHN. Since the clinical manifestation of PPHN usually involves a short onset, genotyping of identified genetic variants in a family with a previous PPHN child could be a sign indicating close monitoring of following children during the perinatal period. Biomarkers currently serve as a valuable tool for early diagnosis, disease staging, they can be used as prognostic indicators of the treatment outcomes, monitoring of treatment progress, and determining therapeutic targets. In general, a biomarker is a biological molecule found in the blood or other body fluids or tissues. In addition, a genetic marker that causes a disease or is associated with susceptibility to disease is a potential biomarker because a circulating protein encoding from a gene can be genetically influenced. An example of a potential use of a genetic biomarker in PAH is the use of a SNP of the COL18A1 gene to predict the outcome of PAH. Lower serum levels of endostatin, which is encoded from COL18A1, have been associated with reduced mortality in PAH (27). A loss-of-function missense variant in the COL18A1 gene, which results in a change in the amino acid composition of endostatin, was linked to lower serum endostatin and was associated with reduced mortality (27). The genotype AG of the rs12483377 SNP had significantly lower serum levels of endostatin compared with the homozygous GG genotype (27). Another example is the use of plasma endothelin-1 as an effective biomarker for monitoring and predicting the outcome of pulmonary hypertension (28,29). In another study, the presence of the rs2070699 T allele of the EDN1 gene that encodes endothelin-1 was associated with higher plasma endothelin-1 levels, which increase the risk of PPHN in neonates with respiratory distress (11). Other potential biomarkers that confer a risk of developing PPHN are plasma tyrosine and phenylalanine (30). A study on polymorphisms of urea cycle enzymes and intermediate metabolites found that 2 intronic SNPs and a synonymous mutation in the CPS1 gene were significantly associated with PPHN (12). The same study found that tyrosine levels were significantly lower and phenylalanine levels were significantly higher in patients with PPHN when compared to controls (12). Other biomarkers of pulmonary diseases, including genetic variants, are extensively reviewed elsewhere (31). Biomarkers may be used for risk stratification of PPHN and as a guide for specific treatments when the evidence is sufficient, especially for at risk newborns. For example, the determination of CPS1 genetic variants and urea cycle intermediate metabolites in newborns with birth asphyxia and meconium-stained amniotic fluid may be a guide for disease vigilance or the use of a selective blockade for endothelin-1 receptor type A to encourage pulmonary vasodilation in newborns with EDN1 genetic variants.
Unlike rare monogenic diseases in which the variants usually reside within coding regions of genes, variants associated with common complex diseases usually reside within non-coding regions (32). Our study examined SNPs in the intron of the WWC2 gene. Intronic SNPs may result in alter RNA splicing. Disrupted RNA splicing results in the loss of exons or the inclusion of introns and an altered protein-coding sequence. Alternative splicing and exon shuffling lead to enhancing the protein diversity. Intron also plays an important role in gene regulation and expression, regulation of mRNA transport, and chromatin assembly (33).
In this study, we were unable to recruit more patients with PPHN in our registry as many former patients were lost to contact and blood samples had not been kept. Therefore, we prospectively studied infants with PPHN together with matching healthy control infants. Since not all neonates with respiratory failure develop PPHN, it seems that PPHN is most likely caused by some type of pulmonary vascular maladaptation precipitated by genetic-environmental interactions. Since environmental factors are a main factor triggering PPHN, genetic studies in patients with idiopathic PPHN might provide powerful insights into the genetics of PPHN. However, recruiting patients with idiopathic PPHN is difficult, as they represent only a small fraction of these patients. In addition, further studies regarding the functional effects of WWC2 gene variants should be carried out to determine their clinical significance in PPHN. The expression of the WWC2 gene could be evaluated either in lung and other tissues obtained from deceased patients with PPHN or in animal models of PPHN, such as mice that have elevated pulmonary arterial pressure and some degree of hypoxemia. Functional studies of the WWC2 gene can be performed by immunohistochemical analysis or western blot using antibodies against the WWC2 protein. WWC2 mRNA expression studies can be performed by various methods such as whole-mount in situ hybridization or quantitative reverse transcriptase polymerase chain reaction (34,35). In this study, we did not perform a functional study of the WWC2 gene since we did not have lung tissue samples obtained from deceased patients with PPHN and a mouse model with PPHN could not be created in our setting.
Conclusions
In conclusion, this is the first study to examine possible associations between genetic variations and PPHN using GWAS across the genome. We identified WWC2 as a possible candidate gene, which may modulate the risk for PPHN. Further studies are required to confirm our findings and to explore the physiological mechanism behind this association.
Acknowledgments
The authors thank Dr. Jakris Eu-ahsunthornwattana of the Department of Community Medicine, Faculty of Medicine Ramathibodi Hospital, Mahidol University for allowing us to use the Thai SNP database. The authors thank Mr. David Patterson of the Office of International Affairs, Faculty of Medicine, Prince of Songkla University for editing the manuscript.
Funding: This study was funded by a grant from the Graduate School and Faculty of Medicine Foundation of Prince of Songkla University, and the Medical Genetics Center, Division of Genomic Medicine and Innovation Support, Department of Medical Sciences, Ministry of Public Health, Thailand.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-280/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-280/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-280/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of Hat Yai Hospital (No. 24/2562) and informed consent was taken from the patients’ parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jain A, McNamara PJ. Persistent pulmonary hypertension of the newborn: Advances in diagnosis and treatment. Semin Fetal Neonatal Med 2015;20:262-71. [Crossref] [PubMed]
- Steurer MA, Jelliffe-Pawlowski LL, Baer RJ, et al. Persistent Pulmonary Hypertension of the Newborn in Late Preterm and Term Infants in California. Pediatrics 2017;139:e20161165. [Crossref] [PubMed]
- Nakwan N, Jain S, Kumar K, et al. An Asian multicenter retrospective study on persistent pulmonary hypertension of the newborn: incidence, etiology, diagnosis, treatment and outcome. J Matern Fetal Neonatal Med 2020;33:2032-7. [Crossref] [PubMed]
- Hernández-Díaz S, Van Marter LJ, Werler MM, et al. Risk factors for persistent pulmonary hypertension of the newborn. Pediatrics 2007;120:e272-82. [Crossref] [PubMed]
- Rocha G, Baptista MJ, Guimarães H. Persistent pulmonary hypertension of non cardiac cause in a neonatal intensive care unit. Pulm Med 2012;2012:818971. [Crossref] [PubMed]
- Bearer C, Emerson RK, O'Riordan MA, et al. Maternal tobacco smoke exposure and persistent pulmonary hypertension of the newborn. Environ Health Perspect 1997;105:202-6. [Crossref] [PubMed]
- Alano MA, Ngougmna E, Ostrea EM Jr, et al. Analysis of nonsteroidal antiinflammatory drugs in meconium and its relation to persistent pulmonary hypertension of the newborn. Pediatrics 2001;107:519-23. [Crossref] [PubMed]
- Steinhorn RH. Diagnosis and treatment of pulmonary hypertension in infancy. Early Hum Dev 2013;89:865-74. [Crossref] [PubMed]
- Pearson DL, Dawling S, Walsh WF, et al. Neonatal pulmonary hypertension--urea-cycle intermediates, nitric oxide production, and carbamoyl-phosphate synthetase function. N Engl J Med 2001;344:1832-8. [Crossref] [PubMed]
- Byers HM, Dagle JM, Klein JM, et al. Variations in CRHR1 are associated with persistent pulmonary hypertension of the newborn. Pediatr Res 2012;71:162-7. [Crossref] [PubMed]
- Mei M, Cheng G, Sun B, et al. EDN1 Gene Variant is Associated with Neonatal Persistent Pulmonary Hypertension. Sci Rep 2016;6:29877. [Crossref] [PubMed]
- Kaluarachchi DC, Smith CJ, Klein JM, et al. Polymorphisms in urea cycle enzyme genes are associated with persistent pulmonary hypertension of the newborn. Pediatr Res 2018;83:142-7. [Crossref] [PubMed]
- Liu X, Mei M, Chen X, et al. Identification of genetic factors underlying persistent pulmonary hypertension of newborns in a cohort of Chinese neonates. Respir Res 2019;20:174. [Crossref] [PubMed]
- Wang M, Zhuang D, Mei M, et al. Frequent mutation of hypoxia-related genes in persistent pulmonary hypertension of the newborn. Respir Res 2020;21:53. [Crossref] [PubMed]
- El-Khazragy N, El Barbary M, Fouad H, et al. Association between genetic variations in carbamoyl-phosphate synthetase gene and persistent neonatal pulmonary hypertension. Eur J Pediatr 2021;180:2831-8. [Crossref] [PubMed]
- Nakwan N, Mahasirimongkol S, Satproedprai N, et al. Possible association between a polymorphism of EPAS1 gene and persistent pulmonary hypertension of the newborn: a case-control study. J Pediatr (Rio J) 2022;98:383-9. [Crossref] [PubMed]
- Germain M, Eyries M, Montani D, et al. Genome-wide association analysis identifies a susceptibility locus for pulmonary arterial hypertension. Nat Genet 2013;45:518-21. [Crossref] [PubMed]
- Kimura M, Tamura Y, Guignabert C, et al. A genome-wide association analysis identifies PDE1A|DNAJC10 locus on chromosome 2 associated with idiopathic pulmonary arterial hypertension in a Japanese population. Oncotarget 2017;8:74917-26. [Crossref] [PubMed]
- Morrell NW, Aldred MA, Chung WK, et al. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J 2019;53:1801899. [Crossref] [PubMed]
- Bendapudi P, Rao GG, Greenough A. Diagnosis and management of persistent pulmonary hypertension of the newborn. Paediatr Respir Rev 2015;16:157-61. [Crossref] [PubMed]
-
GTExPortal - Duong TE, Wu Y, Sos BC, et al. A single-cell regulatory map of postnatal lung alveologenesis in humans and mice. Cell Genom 2022;2:100108. [Crossref] [PubMed]
- Bertero T, Oldham WM, Cottrill KA, et al. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest 2016;126:3313-35. [Crossref] [PubMed]
- Chen YC, Tsai YH, Wang CC, et al. Epigenome-wide association study on asthma and chronic obstructive pulmonary disease overlap reveals aberrant DNA methylations related to clinical phenotypes. Sci Rep 2021;11:5022. [Crossref] [PubMed]
- Puthiyachirakkal M, Mhanna MJ. Pathophysiology, management, and outcome of persistent pulmonary hypertension of the newborn: a clinical review. Front Pediatr 2013;1:23. [Crossref] [PubMed]
- Chida A, Shintani M, Matsushita Y, et al. Mutations of NOTCH3 in childhood pulmonary arterial hypertension. Mol Genet Genomic Med 2014;2:229-39. [Crossref] [PubMed]
- Damico R, Kolb TM, Valera L, et al. Serum endostatin is a genetically determined predictor of survival in pulmonary arterial hypertension. Am J Respir Crit Care Med 2015;191:208-18. [Crossref] [PubMed]
- Yunis A, Nouraie M, Britton N, et al. Endothelin-1 may be a potential biomarker for monitoring changes in nitric oxide metabolism in pulmonary hypertension. J Card Fail 2020;26:S17-8.
- Jankowich MD, Wu WC, Choudhary G. Association of Elevated Plasma Endothelin-1 Levels With Pulmonary Hypertension, Mortality, and Heart Failure in African American Individuals: The Jackson Heart Study. JAMA Cardiol 2016;1:461-9. [Crossref] [PubMed]
- Steurer MA, Oltman S, Baer RJ, et al. Altered metabolites in newborns with persistent pulmonary hypertension. Pediatr Res 2018;84:272-8. [Crossref] [PubMed]
- Jain KK. Biomarkers of pulmonary diseases. In: The Handbook of Biomarkers. New York, NY, USA: Humana Press, 2017:673-88.
- Frayling TM. Genome-wide association studies: the good, the bad and the ugly. Clin Med (Lond) 2014;14:428-31. [Crossref] [PubMed]
- Jo BS, Choi SS. Introns: The Functional Benefits of Introns in Genomes. Genomics Inform 2015;13:112-8. [Crossref] [PubMed]
- Hermann A, Wu G, Nedvetsky PI, et al. The Hippo pathway component Wwc2 is a key regulator of embryonic development and angiogenesis in mice. Cell Death Dis 2021;12:117. [Crossref] [PubMed]
- Höffken V, Hermann A, Pavenstädt H, et al. WWC Proteins: Important Regulators of Hippo Signaling in Cancer. Cancers (Basel) 2021;13:306. [Crossref] [PubMed]


