
Comprehensive genetic analyses of childhood acute leukemia in Iraq using next-generation sequencing
Highlight box
Key findings
• Among Iraqi children with acute leukemia, we disclosed a high frequency of TCF3-PBX1 in ALL, and a frequent AML-M3 subtype, along with recurrent RAS mutations in ALL/AML.
What is known, and what is new?
• Molecular analyses in hematological malignancies provide insights about genetic makeup. Probable etiological factors in leukemogenesis could be disclosed.
• For the first time in Iraq, NGS was performed to disclose the molecular landscape of a cohort of childhood ALL/AML from Iraq in Japan using dried blood spot samples. Our results suggest that the biology of Iraqi childhood acute leukemia is, in part, characteristic, where the war-aftermath environment or geography might play a role.
What is the implication, and what should change now?
• Understanding the biology of acute leukemia in Iraq could help doctors there in modifying the management protocols or arranging the plan for required and applicable analysis in their locations for achieving better results.
Introduction
Childhood acute leukemia has heterogeneous biological and multifactorial etiology mechanisms linked with genetic susceptibility factors and subsequently acquired somatic mutations (1-3). Differences in the incidences, risk factors, and survival of pediatric acute leukemia along with the different frequencies of molecular markers have been reported across various countries (4,5). Such differences could be attributed to the interaction between genomic drivers, which are associated with race and ethnicity, and environmental factors (2,3,6-8).
Over the last four decades of wars and their aftermath in Iraq, the health system underwent a serious regression. No genetic analysis is yet available for diagnosing pediatric acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) in Iraq. Limited diagnostic facilities have a negative impact on disease understanding, management, and consequently its outcome. Meanwhile, childhood leukemia rates doubled over 15 years (1993–2007) according to a study from Basra in Southern Iraq, and the trend was deemed significant when compared to neighboring countries like Kuwait and Oman, as well as the United States (9). Notably, Basra was the nearest spot that experienced repeated gulf wars and was exposed to repeated bombing and by-products of the petroleum fires.
Despite the improved 5-year survival rate of pediatric ALL outcome of more than 90% in developed countries, Iraqi pediatric oncologists struggle to achieve around 70% (10,11). The international collaboration from Japan was, therefore, established aiming to scale up the diagnosis of Iraqi children with acute leukemia by performing molecular analysis using the dried blood spot (DBS) samples; concomitantly, Italy had settled a telemedicine program in the main pediatric oncology center in Baghdad to support in protocol guidance for acute leukemia cases. Through our collaboration studies, the prevalence of RAS mutations was previously noted to be higher among Iraqi childhood ALL and AML than in other countries (12,13). Likewise, acute promyelocytic leukemia (APL) was unusually frequent among Iraqi children with AML in our study (14) and in a report by the Italian team (15).
Next-generation sequencing (NGS) technology with its characteristic high throughput and high sensitivity and specificity provides a good platform for acute leukemia diagnosis and research to improve the understanding of molecular alterations in patients with such diseases. Thus, it aids in refining their treatment plans accordingly. NGS when compared to conventional genetic sequencing, has several advantages such as comprehensive genomic coverage, higher capacity with sample multiplexing, and the ability to sequence hundreds to thousands of genes or gene regions simultaneously (16).
In this new collaborative study, NGS was utilized for the first time for illustrating the landscape of genetic mutations in a series of Iraqi children with ALL and AML, and the DBS-extracted DNA was used for the NGS analysis. We aimed to perform a more comprehensive genetic analysis using NGS and to assess the possible differences in the biology of pediatric acute leukemia in Iraq in association with genetic or non-genetic factors with the consideration of environmental factors. We present this article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-512/rc).
Methods
General information about the study
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The research work was approved by the Ethical Committee of Shinshu University School of Medicine (No. 622/2020), Nagoya University Graduate School of Medicine (No. 18185/2020), and by the Ministry of Health in Iraq (No. 2553/2018). All methods were carried out in accordance with relevant guidelines and regulations and a written informed consent was obtained from all subjects and/or their legal guardian(s).
Five main pediatric oncology centers from Iraq have participated in this study, including Children Welfare Teaching Hospital (CWTH) in Baghdad (the major referral center for childhood cancers in the country), Basra Children’s Specialty Hospital (BCSH) in Basra, Ibn Al-Atheer Hospital for Children (IAH) in Mosul, Hiwa Cancer Hospital (HCH) in Sulaymaniyah, and Jin Pediatric Hematology-Oncology Centre (JPHOC) in Duhok. CWTH, BCSH, and IAH are in Arab provinces, whereas HCH and JPHOC are in Kurdistan, the area inhabited mostly by the Kurdish ethnicity in the north of Iraq. HCH in Sulaymaniyah is the only oncology center in Iraq, which is equipped with hematopoietic stem cell transplantation unit under the supervision of an Italian team, and they are performing the minimal residual disease detection using flow cytometry. Patients in the mentioned centers were treated according to the United Kingdom-Medical Research Council (UK-MRC) protocols for pediatric acute leukemia, including the modified UKALL 2011 for ALL, and AML-MRC15 for AML, described elsewhere (10,17).
Sample collection
In the form of DBS, paired bone marrow (BM) samples were collected at diagnosis (day 0, tumor status), and at (day 30 or 60, remission status), from Iraqi patients aged ≤16 years, who were newly diagnosed with ALL or AML, from June 2016 to December 2019. Providing that no molecular data are available in Iraq upon diagnosis and the samples were received sequentially within the first few weeks of diagnosis, selection bias are not expected. In total, 101 cases were recruited from Iraq, 53 from CWTH, 25 from BCSH, 15 from HCH, and 8 from JPHOC. However, 66 (55 ALL and 11 AML) cases who had paired BM samples (at diagnosis and remission) were eligible for NGS, including 36, 17, 8, and 5, from CWTH, BCSH, HCH, and JPHOC, respectively. The remaining 35 cases were either missing one sample (n=15), died before reaching remission (n=9), insufficient in terms of DNA concentration (n=5), transferred to be treated outside Iraq (n=3), or abandoned therapy (n=3).
Flinders Technology Associates (FTA) paper processing
A few drops of blood from BM aspirate at initial diagnosis and at remission status were applied to the FTA classic card’s filter paper (Cat No. WB120205, GE Healthcare, Buckinghamshire, UK Limited) (18) at the five Iraqi hospitals. After the blood spots were dried for 1 hour at room temperature, the FTA card was kept in a special FTA envelope in a refrigerator for up to several weeks and was then transported by airplane to Japan. Two mm disks (eight disks) were punched out from the DBS on FTA cards using a sterile hole puncher (Harris Micro-Punch, Shunderson Communications Inc., Ottawa, Canada). For the matched remission status samples especially those with hypoplastic BM samples, more DBS disks (up to 40) were consumed to increase the DNA yield. DNA was extracted from the DBS from the samples of FTA cards and was purified using the QIA amp DNA Blood Mini Kit (Cat No. 56304, Qiagen, Ltd., Tokyo, Japan) as per the manufacturer’s instructions. After the extraction, DNA was measured using Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, Life Technologies, MA, USA) as per the manufacturer’s instructions.
Whole-exome sequencing (WES)
NGS analyses have been performed essentially as described (19). Briefly, WES libraries starting from 50–200 ng of DNA have been prepared using a SureSelect Human All Exon V5 bait and SureSelect Reagents (Agilent, Santa Clara, CA, USA) as per the manufacturer’s instructions. The libraries were run on a HiSeq X next-generation sequencer (Illumina, San Diego, CA, USA), with a 2×150-bp paired end-reads option. The sequence reads were aligned to the hg19 reference genome using the Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/) with default parameters and a “–mem” option. Polymerase chain reaction (PCR) duplicates were removed from constructed BAM files using the Picard tools (https://broadinstitute.github.io/picard/).
To identify somatic point mutations, paired tumor-normal data were analyzed using VarScan2. We then called candidate variants in the coding region that have variant allele frequencies (VAF) of >0.1 (in tumor) and <0.05 (in normal), 10 or more reads with the variant, and minor allele frequencies (MAF) of <0.001 in single nucleotide polymorphism (SNP) databases (ESP6500, 1000 genomes, ExAC, and Kaviar). A candidate variant was considered as an artifact and was filtered out if the identical variant was present in 12 irrelevant germline samples with an average VAF of >0.01. The variants were then annotated using ANNOVAR (https://annovar.openbioinformatics.org/).
In total, 50 candidates were randomly selected, and PCR-based deep sequencing was performed. Briefly, a NotI-tagged PCR primers (having 5’-AAGCGGCCGC-3’ tag on their 5’- side) were designed to cover 100–200 bp regions including candidates. PCR products were digested using NotI (New England Biolabs, Ipswich, MA, USA) and concatenated using T4 DNA Ligase (TaKaRa Bio, Otsu, Japan). The concatemers were fragmented to an average length of 400 bp by Covaris M220 (Covaris, Wobam, MA, USA) and were prepared for sequencing using an NEBNext Ultra DNA Prep Kit for Illumina (New England Biolabs) as per the manufacturer’s instructions. BAM files have been assembled, and VAF of candidates has been measured. The candidate is considered present if the VAF in tumor was three times more than that in normal. As a result, (48/50, 96%) were confirmed to be present as somatic mutations.
To identify germline variants, variants with VAF >0.25 in normal data have been picked up using VarScan2. The variants were annotated using ANNOVAR. Genetic diagnoses were considered only when germline variants fulfilled the criteria of “pathogenic” or “likely pathogenic” as provided by the American College of Medical Genetics guideline, as described (20). The zygosity of a variant was considered homozygous when the VAF of the variant exceeded 0.85.
To identify copy number alterations (CNAs), a read count of an exon in a tumor sample was normalized for the total coverage of the sample and was compared with 12 irrelevant germline samples. The exon was considered a candidate of CNAs if the standard deviation of the tumor sample’s read count was >3. If three or more continuous exons are the candidates, the exons are considered affected by amplifications or deletions.
Run of homozygosity (ROH) was identified by detecting a run of homozygous common (>1% MAF in SNP databases) SNPs in normal samples. ROH was classified by simply counting the number of continuous SNPs (10–50 SNPs; short ROH, and >50 SNPs; long ROH). A total of 60 germline samples of Japanese origin were used as control.
Targeted gene sequencing (TGS)
A custom SureSelect bait was designed targeting the whole gene body of 31 genes associated with fusion genes or deletions in B-ALL (Table S1). To detect chromosomal structural variations (SVs), soft-clipped bases were realigned to the hg19 using BLAT (https://hgwdev.gi.ucsc.edu/~kent/src/). A candidate SV supported by five or more reads with the identical breakpoint was visually interrogated using the Integrative Genomics Viewer (https://software.broadinstitute.org/software/igv/).
Whole-genome sequencing (WGS)
A total of 13 cases were analyzed using WGS. WGS libraries were prepared starting from 50–100 ng of DNA using an NEBNext Ultra II DNA Prep Kit for Illumina (New England Biolabs), according to the manufacturer’s instructions. Somatic and germline variants and SVs were detected using the approaches used in WES and TGS. A copy number estimate of 10 kb bin was made simply from the number of reads within the bin divided by the mean coverage of the sample.
Co-occurrence simulation
The probability of co-occurrence of two kinds of genetic alterations was calculated using a Monte-Carlo simulation approach, based on the number of total patients, the number of patients with either of the mutations, and the number of patients with both mutations, as described (21).
Statistics
Statistical analyses were performed using SPSS program v. 28 (SPSS, IBM Corporation, Armonk, NY, USA). The unpaired Student’s t-test was used in determining the significance of differences between two independent groups, and the Mann-Whitney U-test was used for data that were not normally distributed. Chi-square test or Fisher’s exact test was used to compare the frequencies of genetic mutations between our cases and those from other countries. Statistical significance was defined as a P value of <0.05.
Results
Study cohort and design
This study included 49, 6, and 11 cases of B-cell precursor ALL (B-ALL), T-cell precursor ALL (T-ALL), and AML, respectively (Table 1). The median age among B-ALL was 4.2 (1–13) years, with a male to female ratio (M/F) of 1.7, meanwhile, the median age among T-ALL cases was 9.3 (3.5–12.8) years, and 5/6 of them were males. The median white blood cell (WBC) count in B-ALL and T-ALL was 16.4 (2.4–181) ×109/L and 280.5 (4.2–700) ×109/L, respectively. The average age and WBC were significantly higher in T-ALL compared to B-ALL, with P values of 0.007 and <0.001, respectively. The 3-year event-free survival (EFS) was 70.9%, and the 3-year overall survival (OS) was 74.5%. In AML, a higher frequency of APL or French-American-British (FAB) AML-M3 morphology (5/11, 45%) was observed, followed by FAB-M2 (4/11, 36%), and one case of M5 and M6. A case of AML-M2 (UPN49) was found to be secondary to chemotherapy or therapy-related AML (s-AML), for a previously cured germ cell tumor.
Table 1
| Acute leukemia type | Variable | Number of patients (%) |
|---|---|---|
| ALL (n=55) | Sex | |
| Male | 36 (65.5) | |
| Female | 19 (34.5) | |
| Age (years) | ||
| 1–<5 | 29 (52.7) | |
| 5–<10 | 19 (34.6) | |
| ≥10 | 7 (12.7) | |
| WBC (×109/L) | ||
| <20 | 27 (49.1) | |
| 20–<50 | 9 (16.4) | |
| ≥50 | 19 (34.5) | |
| ALL subtypes | ||
| B-ALL | 49 (89.1) | |
| T-ALL | 6 (10.9) | |
| AML (n=11) | Sex | |
| Male | 8 (72.7) | |
| Female | 3 (27.3) | |
| Age (years) | ||
| 1–<5 | 5 (45.5) | |
| 5–<10 | 1 (9.0) | |
| ≥10 | 5 (45.5) | |
| AML subtypes | ||
| M2 | 4 (36.3) | |
| M3 (APL) | 5 (45.5) | |
| M5 | 1 (9.1) | |
| M6 | 1 (9.1) | |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; WBC, white blood cell; APL, acute promyelocytic leukemia.
Either WES (53 cases), or WGS (13 cases) for each patient was performed. Paired tumor (BM specimen at diagnosis) and germline (BM at remission) samples were analyzed for each patient to identify somatic and germline mutations. Additionally, for patients with B-ALL analyzed by WES (40 cases), targeted sequencing was performed to identify fusion genes (Table S1).
Quality assessment of DBS-derived DNA
Since using DBS-derived DNA was unusual for NGS, the performance of our analysis was checked. As a result, an average of 81.0× and 30.3× coverage was obtained in WES and WGS analyses, respectively. The coverage resulted in 97.7% and 97.3% of the coding region covered by 10 or more unique reads, suggesting that DBS-derived DNA can be utilized for NGS.
Somatic mutations
A comprehensive detection of point mutations, small insertions/deletions (indels), copy number variants, and chromosomal SVs was performed in 66 patients with acute leukemia in Iraq (Figure 1, Tables S2,S3).
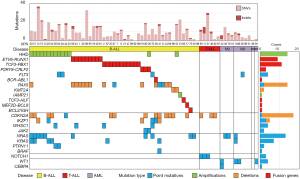
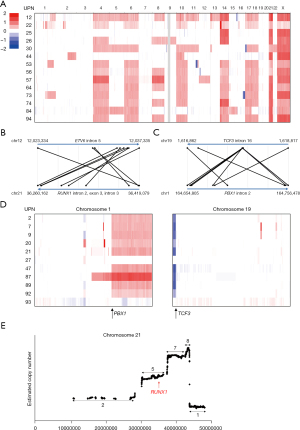
At least one driver mutation in 48 (95%) cases with B-ALL was identified, and accordingly B-ALL cases were classified. In 21 (42%) cases, 2 major subsets of B-ALL, including, high hyperdiploid (HHD) (>50 chromosomes), and ETV6-RUNX1, were identified representing 12, and 9 cases, respectively. The pattern of chromosomal amplification in HHD cases and the positions of chromosomal recombination in ETV6-RUNX1 cases were found to be typical (Figure 2A,2B). Surprisingly, TCF3-PBX1, which usually constitutes 3–5% of a B-ALL cohort, explained 11 (22.4%) cases. This observation was not caused by cross-contamination of samples, because each patient carried a unique chromosomal breakpoint (Figure 2C). Moreover, the fusion gene was frequently associated with the amplification and deletion of chromosomes 1 and 19, respectively (Figure 2D). Four Ph-like ALL cases (3 with P2RY8-CRLF2 and a case with FLT3-tyrosine kinase domain mutation) were identified (22), in addition to a Ph-ALL case with BCR-ABL1. Two of the 3 cases with P2RY8-CRLF2 carried concomitant JAK2 p.Arg683Gly point mutations. Other classifications were PAX5 alterations (four cases) (22), KMT2A (MLL) deletion (two cases), intrachromosomal amplification of chromosome 21 (iAMP21), one case (Figure 2E), TCF3-HLF fusion (one), MEF2D-BCL9 fusion (one), and BCL2-IGH fusion (one case).
Two cases (UPN65 and UPN98) did not carry any mutation associated with B-ALL classification and thus were classified with B-other ALL. The contamination of tumor in the germline sample or the scarcity of tumor cells in the tumor specimen was considered to explain the absence of classification in these two patients (Table S4).
Some mutations showed co-occurrence within a patient. Both HHD and RAS pathway mutations (NRAS, KRAS, PTPN11, and BRAF mutations) were detected in nine patients (P=0.0058) (Table S5). Also, 6/9 patients with ETV6-RUNX1 carried PAX5 mutations (P=0.0066).
In patients with T-ALL or AML, several mutations that are characteristic of these diseases have been identified. T-ALL carried NOTCH1, PTEN, ETV6, IL7R, RUNX1, RPL10, and SUZ12 mutations and CDKN2A deletions. AML carried WT1, CEBPA, FLT3, MYC, KRAS, and NRAS mutations. Because fusion gene detection was not performed in patients who had these diseases and were analyzed by WES, characteristic fusion genes were mostly not identified in these patients; but at least, PML-RARA was detected by WGS in UPN99 with AML-M3 and thus confirmed the PCR result of that patient in Iraq.
The number of somatic point mutations in the coding region was 0–37 (9.9 on average) and was considered comparable with similar diseases in other countries. B-ALL and T-ALL were noted to significantly differ in terms of the number of indels (0.81 vs. 2.66 on average, P=0.002), while the small number of T-ALL cases defies its interpretation. The number of indels looked high in several patients (including UPN11 who carried seven indels); however, the number was not statistically significant.
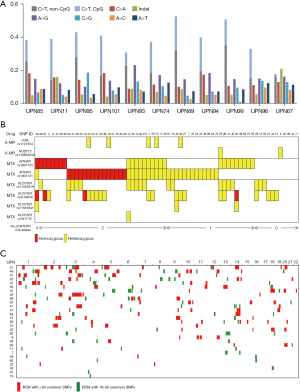
The type of nucleotide alterations of somatic mutations was biased toward C-to-T transitions (40%), suggesting that most somatic point mutations were acquired because of cell division (Figure 3A). Indels accounted for 7.5% of the somatic mutations.
The RAS pathway mutations were present in (16/49, 32.7%) of B-ALL cases and (3/11, 27.3%) of AML cases. Thus, in line with our previous reports, RAS mutations are prevalent among Iraqi children with acute leukemia compared with that of other countries (23-28). Three patients carried 2 RAS pathway mutations in ALL (NRAS and KRAS in both UPN30 and UPN57, KRAS and BRAF in UPN41), and one AML case (UPN10) had NRAS and KRAS mutations.
Germline variations
Germline mutations were analyzed; however, those related to the known inherited diseases were not identified. We also could not point out any pathogenic variants associated with leukemia predisposition. Meanwhile, several drug metabolism-associated SNPs were disclosed for 6-mercaptopurine (6-MP) and methotrexate (MTX) (29-31) (Figure 3B). Two SNPs (ITPA rs1127354 and NUDT15 rs116855232) associated with 6-MP toxicity were present with MAF of 0.038, and 0.015, respectively. MTHFR rs1801131 (MTHFR-A or c.1298A>C) and rs1801133 (MTHFR-C or c.677C>T), which affect MTX metabolism, were frequent (MAF of 0.409 and 0.265, respectively). As a result, (23/66, 34.8%) cases of our cohort were affected by 2 or more MTHFR-A/C risk alleles. SLCO1B1, another MTX catalyzer, carried several drug metabolism-associated SNPs including rs11045819 (MAF =0.144) and rs4149056 (MAF =0.197). Additionally, it may be notable that SLCO1B1 was also affected by rare nonsense mutations including rs7158941 (p.Arg580Ter, three patients, MAF =0.023) and p.Trp171Ter (one patient).
ROH was frequently observed in the germline of children with acute leukemia in Iraq, possibly because of consanguinity (Figure 3C). In total, (29/66, 43.9%) patients carried at least 1 ROH that had a length of >10 Mb. However, a significant accumulation of ROH could not be identified. At the very least, the lengths of ROH were significantly longer compared with those of Japanese samples [Iraq: 0–392,289,752 bp (70,539,478 bp on average); Japan: 0–148,869,110 bp (8,397,369 bp on average), P=2.6×10−6].
Genetic findings and clinical presentations
Several fusion genes were associated with clinical parameters; 9 patients with ETV6-RUNX1 fusion gene had a median age of 4 (3–4.75) years, with M/F ratio of 3.5, and a median WBC of 11.7 (4.5–72) ×109/L (Table 2). The average WBC associated with ETV6-RUNX1 cases was lower than those B-ALL cases without ETV6-RUNX1; however, it was of no significance (22.5 vs. 42) ×109/L, respectively (P=0.237). Eleven patients with TCF3-PBX1 fusion gene had a median age of 5.7 (2–12) years, M/F ratio of 4.5, and a median WBC of 52.5 (4.6–152) ×109/L. The average WBC in patients with TCF3-PBX1 was significantly higher than those TCF3-PBX1-negative B-ALL cases (63.4 vs. 31.2) ×109/L, respectively (P=0.033), whereas the average number of somatic mutations per patient associated with TCF3-PBX1 was significantly lower than those B-ALL cases without TCF3-PBX1 (5.6 vs. 11.9), respectively (P=0.023).
Table 2
| UPN/sex | Modified UKALL-2011 clinical-based risk factors classification | Genetic aberrations | Notes and outcome**** | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | Initial WBC ×109/L | Initial CSF (CNS status)* | Pre-phase steroid response** | Post-induction BMA status | Protocol regimen | ALL classification | Potentially deleterious somatic point mutation | Relapse, site/time in weeks | Death, time of death in weeks/cause | Cured, in continuous CR and others | |||
| 1/M | 4.20 | 34.07 | – | Good | CR by Morph*** | A | ETV6-RUNX1 | CNS/54 | Follow-up | ||||
| 19/M | 3.50 | 11.6 | – | Good | CR by Morph | A | ETV6-RUNX1 | Follow-up | |||||
| 43/M | 4.10 | 22.4 | – | Good | CR by Morph | A | ETV6-RUNX1 | WHSC1 | On therapy | ||||
| 58/M | 4.00 | 5.1 | – | Good | CR by Morph | A | ETV6-RUNX1 | NRAS | On therapy | ||||
| 61/F | 3.00 | 11.4 | – | Good | CR by Morph | A | ETV6-RUNX1 | Follow-up | |||||
| 62/M | 3.00 | 29.8 | – | NU | Flow-MRD: 0.005% | A | ETV6-RUNX1 | On therapy | |||||
| 68/M | 4.00 | 4.5 | – | NU | Flow-MRD: 0.001% | A | ETV6-RUNX1 | On therapy | |||||
| 95/F | 4.75 | 11.7 | – | Good | CR by Morph | A | ETV6-RUNX1 | On therapy | |||||
| 40/F | 3.75 | 72 | – | Good | CR by Morph | B | ETV6-RUNX1 | U2AF1 | Follow-up | ||||
| 44/F | 6.10 | 5.2 | – | Good | CR by Morph | A | HHD | CDKN2A, CDKN2A, CHD4 | Follow-up | ||||
| 64/F | 4.00 | 7.4 | – | NU | Flow-MRD: 0.003% | A | HHD | KRAS | Follow-up | ||||
| 56/F | 6.00 | 2.4 | – | Good | CR by Morph | A | HHD | PTPN11, WHSC1 | Follow-up | ||||
| 57/M | 2.50 | 11.1 | – | Good | CR by Morph | A | HHD | KRAS, NRAS | On therapy | ||||
| 73/M | 4.70 | 5.94 | – | Good | CR by Morph | A | HHD | NRAS, IKZF3 | On therapy | ||||
| 74/F | 6.50 | 8.32 | – | Good | CR by Morph | A | HHD | NRAS | Follow-up | ||||
| 84/F | 5.10 | 16.4 | – | NU | CR by Morph | A | HHD | PTPN11, FLT3, ARID5B | Follow-up | ||||
| 94/F | 2.75 | 12.63 | – | Good | CR by Morph | A | HHD | FLT3 | On therapy | ||||
| 22/F | 1.10 | 9.6 | – | Good | CR by Morph | A | HHD | 84/infection | |||||
| 25/F | 5.60 | 5.7 | – | Good | CR by Morph | A | HHD | IKZF1, KRAS | BM/111 | 114/PD | |||
| 30/M | 7.00 | 71.5 | CNS3 | Good | CR by Morph | B | HHD | KRAS, NRAS | BM/166 | 170/PD | |||
| 12/M | 10.70 | 15.8 | – | Poor | CR by Morph | B | HHD | NRAS, KMT2D, KMT2D | Follow-up | ||||
| 2/M | 5.70 | 33.7 | – | Good | CR by Morph | A | TCF3-PBX1 | WHSC1 | Follow-up | ||||
| 21/F | 7.60 | 16.6 | – | Good | CR by Morph | A | TCF3-PBX1 | SETD2 | Abandon/12 weeks | ||||
| 92/M | 2.00 | 4.6 | – | Good | CR by Morph | A | TCF3-PBX1 | CNS/37 | On palliative therapy | ||||
| 7/M | 7.00 | 40.7 | – | Poor | CR by Morph | B | TCF3-PBX1 | Follow-up | |||||
| 87/M | 12.00 | 6.21 | – | Good | CR by Morph | B | TCF3-PBX1 | KRAS | On therapy | ||||
| 9/M | 4.20 | 134 | – | Good | Not in CR by Morph | C | TCF3-PBX1 | Follow-up | |||||
| 89/F | 10.00 | 65.75 | – | Good | CR by Morph | B | TCF3-PBX1 | Follow-up | |||||
| 93/M | 9.90 | 52.5 | – | Poor | CR by Morph | B | TCF3-PBX1 | On therapy | |||||
| 20/M | 3.40 | 125 | – | Good | CR by Morph | B | TCF3-PBX1 | Follow-up | |||||
| 27/M | 2.20 | 152 | – | Good | CR by Morph | B | TCF3-PBX1 | PAX5 | Follow-up | ||||
| 47/M | 3.00 | 66.14 | – | Good | CR by Morph | B | TCF3-PBX1 | IKZF3 | CNS/63 | 77/infection | |||
| 24/M | 2.20 | 14.6 | – | Good | CR by Morph | A | PAX5alt | TP53 | On therapy | ||||
| 69/M | 3.90 | 89.19 | – | Good | CR by Morph | B | PAX5alt | On therapy | |||||
| 41/F | 13.00 | 11.8 | – | Good | CR by Morph | B | PAX5alt | KRAS, BRAF, PAX5, XBP1 | 45/infection | ||||
| 26/M | 2.00 | 14.4 | – | Good | CR by Morph | A | P2RY8-CRLF2 | JAK2 | On therapy | ||||
| 80/M | 3.00 | 47.4 | – | NU | Flow-MRD: 0.001% | A | P2RY8-CRLF2 | NRAS, JAK2 | On therapy | ||||
| 101/M | 4.20 | 85.07 | – | Good | CR by Morph | B | P2RY8-CRLF2 | On therapy | |||||
| 70/M | 7.40 | 46.5 | – | Good | CR by Morph | A | Ph-like (FLT3) | NRAS, IKZF1, FLT3, WHSC1 | BM + CNS/61 | 92/PD | |||
| 53/F | 2.90 | 3.8 | – | Good | CR by Morph | A | del(11)(q23) | Follow-up | |||||
| 96/M | 3.25 | 23 | – | Poor | CR by Morph | B | del(11)(q23) | On therapy | |||||
| 28/M | 6.80 | 144 | CNS3 | Good | CR by Morph | B | BCR-ABL1 | IKZF1 | Follow-up | ||||
| 75/F | 7.75 | 3.7 | – | Good | CR by Morph | A | BCL2-IGH | NRAS | CNS/111 | 112/PD | |||
| 66/F | 13.00 | 4 | – | NU | Flow-MRD: 0.009% | B | TCF3-HLF | BM/101 | 102/PD | ||||
| 13/M | 3.60 | 58.3 | – | Good | CR by Morph | B | iAMP21 | BCORL1, CSF3R | BM/118 | 149/PD | |||
| 17/M | 7.40 | 30.15 | – | Good | CR by Morph | A | MEF2D-BCL9 | ARID1A | BM + CNS/47 | 113/PD | |||
| 8/F | 7.11 | 181 | CNS3 | Good | CR by Morph | B | B-other ALL | PAX5 | Follow-up | ||||
| 65/M | 1.00 | 51 | – | NU | Flow-MRD: 0.02% | B | B-other ALL | NRAS | On therapy | ||||
| 98/M | 8.00 | 3.16 | – | Poor | Not in CR by Morph | C | B-other ALL | 24/PD | |||||
| 11/M | 9.20 | 73 | – | Poor | CR by Morph | B | T-ALL | NOTCH1, ETV6, IL7R | Follow-up | ||||
| 46/M | 3.50 | 563 | – | Poor | CR by Morph | B | T-ALL | PTEN | 43/infection | ||||
| 67/F | 8.00 | 4.2 | – | NU | Flow-MRD: 0.001% | B | T-ALL | Follow-up | |||||
| 82/M | 10.60 | 700 | CNS3 | NU | CR by Morph | B | T-ALL | CNS/45 | 53/PD | ||||
| 85/M | 12.80 | 111 | – | Good | CR by Morph | B | T-ALL | NOTCH1, RPL10 | Follow-up | ||||
| 31/M | 9.40 | 450 | – | Good | CR by Morph | B | T-ALL | NOTCH1, RUNX1, SUZ12, SUZ12 | On therapy | ||||
*, CNS status, CNS3 defined as CSF of >5 WBC/µL and cytospin positive for blasts; **, a seven-day pre-phase steroid response defined as a drop in peripheral blasts count of <1.0×109/L at day 8; ***, CR by Morph, BM blasts <5% with normal hematopoietic recovery; ****, no patient had testicular involvement at initial diagnosis, and no case in our cohort had Down syndrome. ALL, acute lymphoblastic leukemia; UPN, unique patient number; WBC, white blood cells; CSF, cerebrospinal fluid; CNS, central nervous system; BMA, bone marrow aspirate; CR, complete remission; M, male; Morph, morphology; F, female; NU, not used; Flow-MRD, flow cytometry based-minimal residual disease; HHD, high hyperdiploidy; BM, bone marrow; PD, progressive disease.
Several drug metabolism-associated SNPs were found to be correlated with adverse effects of chemotherapy (Table 3); UPN93, who carried three MTHFR risk alleles (heterozygous MTHFR-A and homozygous MTHFR-C), had experienced frequent interruptions in chemotherapy protocol owing to neutropenia or elevated liver function test. Likewise, UPN25, who carried heterozygous NUDT15 and MTHFR-C risk alleles, had recurrent febrile neutropenia and abnormal liver function test, resulting in relapse, and eventually died. Additionally, UPN87, who carried homozygous MTHFR-C, suffered from frequent neutropenia. Finally, UPN80 with heterozygous MTHFR-C and UPN64 with homozygous MTHFR-A had been complaining from MTX toxicity, with delay in their treatment progress.
Table 3
| UPN/sex | Age (years) | Drug metabolism-associated SNPs | Clinical notes of drug toxicity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methotrexate | 6-mercaptopurine | ||||||||||||
| SLCO1B1 | MTHFR | ITPA | NUDT15 | ||||||||||
| rs4149056 | rs11045819 | rs71581941 | p.W171X | rs1801131 | rs1801133 | rs1127354 | rs116855232 | ||||||
| 1/M | 4.20 | het | – | – | – | het | het | het | – | ||||
| 19/M | 3.50 | – | – | – | – | het | – | – | – | ||||
| 43/M | 4.10 | – | – | – | – | het | – | – | – | ||||
| 58/M | 4.00 | – | het | – | – | het | het | – | – | ||||
| 61/F | 3.00 | – | het | – | – | het | – | – | – | ||||
| 62/M | 3.00 | – | het | – | – | – | – | – | – | ||||
| 68/M | 4.00 | – | het | – | – | – | hom | – | – | ||||
| 95/F | 4.75 | het | – | – | – | hom | – | – | – | ||||
| 40/F | 3.75 | – | – | – | – | – | – | – | – | ||||
| 44/F | 6.10 | het | – | – | – | – | – | – | – | ||||
| 64/F | 4.00 | het | – | – | – | hom | – | – | – | MTX toxicity | |||
| 56/F | 6.00 | – | het | – | – | hom | – | – | – | ||||
| 57/M | 2.50 | – | – | – | – | hom | – | – | – | ||||
| 73/M | 4.70 | – | – | – | – | het | het | het | – | ||||
| 74/F | 6.50 | – | het | – | – | het | het | – | – | ||||
| 84/F | 5.10 | – | – | – | – | het | – | – | – | ||||
| 94/F | 2.75 | – | het | – | – | het | het | – | – | ||||
| 22/F | 1.10 | het | – | – | – | – | – | – | – | ||||
| 25/F | 5.60 | het | het | – | – | – | het | – | het | Frequent FN, abnormal LFT and jaundice | |||
| 30/M | 7.00 | – | – | – | – | het | het | – | – | ||||
| 12/M | 10.70 | – | het | – | het | het | het | – | – | ||||
| 2/M | 5.70 | het | – | – | – | – | hom | – | – | ||||
| 21/F | 7.60 | het | – | – | – | hom | – | – | – | ||||
| 92/M | 2.00 | – | – | – | – | – | het | – | – | ||||
| 7/M | 7.00 | – | – | – | – | het | – | – | – | ||||
| 87/M | 12.00 | hom | – | – | – | – | hom | – | – | Recurrent neutropenia | |||
| 9/M | 4.20 | – | – | – | – | – | hom | – | – | ||||
| 89/F | 10.00 | – | – | – | – | hom | – | – | – | ||||
| 93/M | 9.90 | hom | – | het | – | het | hom | – | – | Recurrent neutropenia/and elevated LFT | |||
| 20/M | 3.40 | het | – | – | – | hom | – | – | – | ||||
| 27/M | 2.20 | het | – | – | – | het | – | – | – | ||||
| 47/M | 3.00 | – | – | – | – | – | hom | – | – | ||||
| 24/M | 2.20 | – | – | – | – | het | – | – | – | ||||
| 69/M | 3.90 | – | het | – | – | hom | – | – | – | ||||
| 41/F | 13.00 | – | het | – | – | – | het | – | – | ||||
| 26/M | 2.00 | het | – | – | – | het | – | het | – | ||||
| 80/M | 3.00 | hom | – | het | – | – | het | – | – | MTX toxicity | |||
| 101/M | 4.20 | – | het | – | – | het | het | – | – | ||||
| 70/M | 7.40 | – | het | – | – | het | – | – | – | ||||
| 53/F | 2.90 | het | – | – | – | hom | – | het | – | ||||
| 96/M | 3.25 | het | het | – | – | – | het | – | – | ||||
| 28/M | 6.80 | het | het | – | – | – | – | – | – | ||||
| 75/F | 7.75 | – | – | – | – | hom | – | – | – | ||||
| 66/F | 13.00 | – | – | – | – | het | – | – | – | MTX toxicity? Frequent neutropenia | |||
| 13/M | 3.60 | – | het | – | – | – | – | – | – | ||||
| 17/M | 7.40 | – | – | – | – | het | het | – | – | ||||
| 8/F | 7.11 | – | – | – | – | – | – | – | het | ||||
| 65/M | 1.00 | – | – | – | – | – | hom | – | – | ||||
| 98/M | 8.00 | – | – | – | – | hom | – | – | – | ||||
| 11/M | 9.20 | – | – | – | – | – | – | – | – | ||||
| 46/M | 3.50 | – | het | – | – | – | het | – | – | ||||
| 67/F | 8.00 | – | – | – | – | – | het | – | – | ||||
| 82/M | 10.60 | – | – | – | – | – | het | – | – | ||||
| 85/M | 12.80 | het | het | – | – | hom | – | – | – | ||||
| 31/M | 9.40 | – | het | – | – | hom | – | – | – | ||||
SNPs, single nucleotide polymorphism; ALL, acute lymphoblastic leukemia; UPN, unique patient number; M, male; het, heterozygous; F, female; hom, homozygous; MTX, methotrexate; FN, febrile neutropenia; LFT, liver function test.
Discussion
The use of FTA cards for conventional molecular analysis including PCR and Sanger sequencing for Iraqi pediatric acute leukemia was previously reported (12-14,18). Whereas in this study, and for the first time NGS was utilized for illustrating the landscape of genetic mutations in a series of Iraqi children with acute leukemia.
Our results disclosed apparent differences in some genetic aberrations, including the unusually high frequency of TCF3-PBX1 fusion gene in ALL (22.4%) and the prevalent APL in AML (45.5%), along with the high frequency of RAS signaling pathway mutations in both ALL (38.8%) and AML (36.4%). Less frequent, however, still comparable results were detected with ETV6-RUNX1 (18.4%), PAX5alt (6.1%), and Ph-like ALL (8.2%) compared to those in the developed world. While HHD (24.5%), BCR-ABL1 (2%), iAMP21 (2%), KMT2A (MLL) deletion (2%), MEF2D-BCL9 (2%), and TCF3-HLF (2%), were similar to those in other studies (19,22,32).
Risk stratification of our cohort according to the clinical characteristics set in the modified UKALL-11 protocol (10) assigned (28/55, 51%) as good risk group eligible for regimen-A treatment plan. Although a total of (32/55, 58.2%) cases carried the favorable risk according to genetic subsets made of ETV6-RUNX1, HHD, and TCF3-PBX1, the stepwise risk refinements, by combining the data, had recognized only (20/55, 36.4%), with the favorable prognostic criteria. Among them, (18/20, 90%) were in continuous complete remission, whether finished or still under treatment, albeit 1 died from infection before completing the therapy. As a result, the 3-year EFS was 70.9%, and the 3-year OS was 74.5%.
One of the striking observations of this study is the unprecedented high frequency of B-ALL cases possessing TCF3-PBX1 fusion gene associated with the translocation t(1;19)(q23;p13), (11/49, 22.4%). Abundance of TCF3-PBX1 in the current cohort is significantly higher than several studies from different ethnicities and countries, ranging from 3% to 7.2% (33-37), including our previous report of (11/264, 4.2%) in pediatric ALL in Iraq (18). Arguably, there might be an underestimation of the frequency of TCF3-PBX1 using the DBS-derived RNA in our previous study compared to DNA. Also, in the previous study the cases were not defined whether of B or T-ALL subtype. Notably, although the number of cases in this study is fewer, our current results are supported both by the chromosomal breakpoints and copy number changes of chromosomes (1 and 19). In fact, our frequency was significantly higher than neighboring Arab countries; Saudi Arabia of 3.4% (38), and Palestine of 7.3% (39), as well as than the Middle Eastern countries of 6.2% (40). Remarkably, TCF3-PBX1 incidence in this report is higher even than that of the African-American B-ALL cases of 16.3% and the Mexican of 14.6% (4,33).
Among AML cases, APL subtype was recurrent (5/11, 45.5%) representing about half of our AML cohort. Of note, frequencies of (9/26, 35%) and (24/134, 18%) of APL among Iraqi children with AML were reported by Testi et al., and Al-Kzayer et al. (based on molecular diagnosis), respectively (14,15). Interestingly, APL seems to be a prevalent AML subtype in Iraq, and records from adolescents and adults in locally published study over 5-year period in a single center at Sulaymaniyah province by Tawfiq et al. (41) showed that APL represented 25.5% of the total AML cases. Indeed, compared to nearby Middle Eastern countries, our frequency is yet higher than that in Saudi Arabia, Israel, Oman, and Iran, which is 3.4%, 8%, 13%, and 16%, respectively (5). Our incidence was also higher than Japan (9%) and other international registries including the United States (5–10%) and Switzerland (2%) (5).
In agreement with our previous work (12,13), in B-ALL, the most frequent somatic mutations were those in the RAS signaling pathway made up of 10 NRAS, 6 KRAS, 2 PTPN11, and 1 BRAF, which were detected in (16/49, 32.7%), including 3 with double mutations. Compared to literature, the overall somatic RAS signaling pathway mutations of around 39% in Iraqi children with ALL are among the highest reported frequencies. Our incidence was comparable to that reported by Case et al. (42) with overall mutations of (26/86, 30%) of childhood ALL cases, provided that FLT3 mutations were excluded from their results. Moreover, our frequency was higher than those reported by Liang et al. (24), with overall RAS mutations of (122/530, 23%) of B-ALL Taiwanese pediatric cohort (P=0.16), and by Zhang et al. (23), with overall mutations of (24/114, 21.1%) in 23 Chinese children with B-ALL (P=0.1). RAS mutations were reported in less frequency of 15–20% in previous studies among childhood ALL (25-28). Wiemels et al. elucidated that RAS mutation frequency among Hispanics was > twice compared to non-Hispanic whites, of 28%, and 13%, respectively, in their cohort, and that HHD-associated RAS mutations were 30%; while, in our series, the latter was 75% (28).
Numerous researchers had investigated the role of environmental exposure to chemicals, including hydrocarbons, and the risk factors behind childhood acute leukemia. Moreover, RAS oncogene was linked to hydrocarbons and other environmental insults. However, whether such association is causal in fact or not remains unclear (7,12,25,28,43).
Iraq was exposed to environmental and chemical hazards that carried potential health risks during repeated wars. Furthermore, the chaotic situation that characterized Iraq, as a consequence of repeated wars, and the damaged infrastructures had resulted in an ongoing process of undifferentiated water and air pollution, with a negative impact on several health aspects in Iraq, including cancer (9,11,12,25,44).
Although racial, ethnic, and geographic differences in the frequency of molecular markers of childhood ALL are widely of concern, the distinct biological difference in the genetics of Iraqi childhood acute leukemia, which varies even with the surrounding countries and sometimes in a significant manner, despite the ethnic similarity, may emphasize the concept of the environmental impact, especially considering Iraq has a war zone environment.
Conclusions
Apart from disclosing the high frequency of TCF3-PBX1, NGS confirmed our previous finding of recurrent RAS mutations in Iraqi childhood acute leukemia. Our results suggest that the biology of Iraqi childhood acute leukemia is in part characteristic, where the war-aftermath environment or geography might play a role. Given the environmental differences in respect to the above complicated status of Iraq, our findings maybe of special interest to encourage more studies enrolling more ALL/AML cases from Iraq to focus on this issue.
Acknowledgments
We thank Mrs. Sadako Kamiya and the non-governmental organization “Japan Chernobyl Foundation” (JCF) for supporting this study. Furthermore, we thank all collaborators from different hospitals in Iraq who have made this work possible.
Funding: This work was supported by a grant from the “Japan Society for the Promotion of Science” (JSPS) for the Grants-in-Aid for Scientific Research (KAKENHI) [18K07295 to LFY Al-Kzayer and Y Okuno], through Shinshu University School of Medicine, Nagano, Matsumoto, Japan. In addition to the grant from “TERUMO LIFE SCIENCE FOUNDATION” to [LFY Al-Kzayer and Y Okuno], for our overseas research in 2019, in Japan. We are thankful to the above agencies for their generous grants.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-512/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-512/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-512/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The research work was approved by the Ethical Committee of Shinshu University School of Medicine (No. 622/2020), Nagoya University Graduate School of Medicine (No. 18185/2020), and by the Ministry of Health in Iraq (No. 2553/2018). All methods were carried out in accordance with relevant guidelines and regulations and informed consent was obtained from all subjects and/or their legal guardian(s).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greaves M. Author Correction: A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer 2018;18:526. [Crossref] [PubMed]
- Buffler PA, Kwan ML, Reynolds P, et al. Environmental and genetic risk factors for childhood leukemia: appraising the evidence. Cancer Invest 2005;23:60-75. [Crossref] [PubMed]
- Hein D, Borkhardt A, Fischer U. Insights into the prenatal origin of childhood acute lymphoblastic leukemia. Cancer Metastasis Rev 2020;39:161-71. [Crossref] [PubMed]
- Jiménez-Morales S, Miranda-Peralta E, Saldaña-Alvarez Y, et al. BCR-ABL, ETV6-RUNX1 and E2A-PBX1: prevalence of the most common acute lymphoblastic leukemia fusion genes in Mexican patients. Leuk Res 2008;32:1518-22. [Crossref] [PubMed]
- Zhang L, Samad A, Pombo-de-Oliveira MS, et al. Global characteristics of childhood acute promyelocytic leukemia. Blood Rev 2015;29:101-25. [Crossref] [PubMed]
- Rodriguez-Galindo C, Friedrich P, Alcasabas P, et al. Toward the Cure of All Children With Cancer Through Collaborative Efforts: Pediatric Oncology As a Global Challenge. J Clin Oncol 2015;33:3065-73. [Crossref] [PubMed]
- Lim JY, Bhatia S, Robison LL, et al. Genomics of racial and ethnic disparities in childhood acute lymphoblastic leukemia. Cancer 2014;120:955-62. [Crossref] [PubMed]
- Chen W, Liu D, Wang G, et al. Screening diagnostic markers for acute myeloid leukemia based on bioinformatics analysis. Transl Cancer Res 2022;11:1722-9. [Crossref] [PubMed]
- Hagopian A, Lafta R, Hassan J, et al. Trends in childhood leukemia in Basrah, Iraq, 1993-2007. Am J Public Health 2010;100:1081-7. [Crossref] [PubMed]
- Al-Hadad SA, Al-Jadiry MF, Ghali HH, et al. Treatment of childhood acute lymphoblastic leukemia in Iraq: a 17-year experience from a single center. Leuk Lymphoma 2021;62:3430-9. [Crossref] [PubMed]
- Al-Hadad SA, Al-Jadiry MF, Al-Darraji AF, et al. Reality of pediatric cancer in Iraq. J Pediatr Hematol Oncol 2011;33:S154-6. [Crossref] [PubMed]
- Al-Kzayer LF, Sakashita K, Al-Jadiry MF, et al. Analysis of KRAS and NRAS Gene Mutations in Arab Asian Children With Acute Leukemia: High Frequency of RAS Mutations in Acute Lymphoblastic Leukemia. Pediatr Blood Cancer 2015;62:2157-61. [Crossref] [PubMed]
- Al-Kzayer LF, Sakashita K, Al-Jadiry MF, et al. Frequent coexistence of RAS mutations in RUNX1-mutated acute myeloid leukemia in Arab Asian children. Pediatr Blood Cancer 2014;61:1980-5. [Crossref] [PubMed]
- Al-Kzayer LF. Analysis of class I and II aberrations in Iraqi childhood acute myeloid leukemia using filter paper cards. Ann Hematol 2014;93:949-55. [Crossref] [PubMed]
- Testi AM, Al-Hadad SA, Al-Jadiry MF, et al. Impact of international collaboration on the prognosis of childhood acute promyelocytic leukemia in Iraq. Haematologica 2006;91:509-12. [PubMed]
- Zhang HH, Wang HS, Qian XW, et al. Ras pathway mutation feature in the same individuals at diagnosis and relapse of childhood acute lymphoblastic leukemia. Transl Pediatr 2020;9:4-12. [Crossref] [PubMed]
- Burnett AK, Russell NH, Hills RK, et al. Optimization of chemotherapy for younger patients with acute myeloid leukemia: results of the medical research council AML15 trial. J Clin Oncol 2013;31:3360-8. [Crossref] [PubMed]
- Al-Kzayer LF, Sakashita K, Matsuda K, et al. Genetic evaluation of childhood acute lymphoblastic leukemia in Iraq using FTA cards. Pediatr Blood Cancer 2012;59:461-7. [Crossref] [PubMed]
- Suzuki K, Okuno Y, Kawashima N, et al. MEF2D-BCL9 Fusion Gene Is Associated With High-Risk Acute B-Cell Precursor Lymphoblastic Leukemia in Adolescents. J Clin Oncol 2016;34:3451-9. [Crossref] [PubMed]
- Muramatsu H, Okuno Y, Yoshida K, et al. Clinical utility of next-generation sequencing for inherited bone marrow failure syndromes. Genet Med 2017;19:796-802. [Crossref] [PubMed]
- Murakami N, Okuno Y, Yoshida K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood 2018;131:1576-86. [Crossref] [PubMed]
- Ratti S, Lonetti A, Follo MY, et al. B-ALL Complexity: Is Targeted Therapy Still A Valuable Approach for Pediatric Patients? Cancers (Basel) 2020;12:3498. [Crossref] [PubMed]
- Zhang H, Wang H, Qian X, et al. Genetic mutational analysis of pediatric acute lymphoblastic leukemia from a single center in China using exon sequencing. BMC Cancer 2020;20:211. [Crossref] [PubMed]
- Liang DC, Chen SH, Liu HC, et al. Mutational status of NRAS, KRAS, and PTPN11 genes is associated with genetic/cytogenetic features in children with B-precursor acute lymphoblastic leukemia. Pediatr Blood Cancer 2018;65: [Crossref] [PubMed]
- Shu XO, Perentesis JP, Wen W, et al. Parental exposure to medications and hydrocarbons and ras mutations in children with acute lymphoblastic leukemia: a report from the Children's Oncology Group. Cancer Epidemiol Biomarkers Prev 2004;13:1230-5. [Crossref] [PubMed]
- Perentesis JP, Bhatia S, Boyle E, et al. RAS oncogene mutations and outcome of therapy for childhood acute lymphoblastic leukemia. Leukemia 2004;18:685-92. [Crossref] [PubMed]
- Yamamoto T, Isomura M, Xu Y, et al. PTPN11, RAS and FLT3 mutations in childhood acute lymphoblastic leukemia. Leuk Res 2006;30:1085-9. [Crossref] [PubMed]
- Wiemels JL, Zhang Y, Chang J, et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia 2005;19:415-9. [Crossref] [PubMed]
- Zhou H, Li L, Yang P, et al. Optimal predictor for 6-mercaptopurine intolerance in Chinese children with acute lymphoblastic leukemia: NUDT15, TPMT, or ITPA genetic variants? BMC Cancer 2018;18:516. [Crossref] [PubMed]
- Fukushima H, Fukushima T, Sakai A, et al. Polymorphisms of MTHFR Associated with Higher Relapse/Death Ratio and Delayed Weekly MTX Administration in Pediatric Lymphoid Malignancies. Leuk Res Treatment 2013;2013:238528. [Crossref] [PubMed]
- Treviño LR, Shimasaki N, Yang W, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol 2009;27:5972-8. [Crossref] [PubMed]
- Inaba H, Mullighan CG. Pediatric acute lymphoblastic leukemia. Haematologica 2020;105:2524-39. [Crossref] [PubMed]
- Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA 2003;290:2001-7. [Crossref] [PubMed]
- Lin A, Cheng FWT, Chiang AKS, et al. Excellent outcome of acute lymphoblastic leukaemia with TCF3-PBX1 rearrangement in Hong Kong. Pediatr Blood Cancer 2018;65:e27346. [Crossref] [PubMed]
- Yen HJ, Chen SH, Chang TY, et al. Pediatric acute lymphoblastic leukemia with t(1;19)/TCF3-PBX1 in Taiwan. Pediatr Blood Cancer 2017;64: [Crossref] [PubMed]
- Wang Y, Xue YJ, Lu AD, et al. Long-Term Results of the Risk-Stratified Treatment of TCF3-PBX1-Positive Pediatric Acute Lymphoblastic Leukemia in China. Clin Lymphoma Myeloma Leuk 2021;21:e137-44. [Crossref] [PubMed]
- Asai D, Imamura T, Yamashita Y, et al. Outcome of TCF3-PBX1 positive pediatric acute lymphoblastic leukemia patients in Japan: a collaborative study of Japan Association of Childhood Leukemia Study (JACLS) and Children's Cancer and Leukemia Study Group (CCLSG). Cancer Med 2014;3:623-31. [Crossref] [PubMed]
- Ahmed AM, Al-Trabolsi H, Bayoumy M, et al. Improved Outcomes of Childhood Acute Lymphoblastic Leukemia: A Retrospective Single Center Study in Saudi Arabia. Asian Pac J Cancer Prev 2019;20:3391-8. [Crossref] [PubMed]
- Shawahna R, Mosleh S, Odeh Y, et al. Clinical characteristics and outcomes of patients with pediatric acute lymphoblastic leukemia after induction of chemotherapy: a pilot descriptive correlational study from Palestine. BMC Res Notes 2021;14:259. [Crossref] [PubMed]
- Al-Mulla NA, Chandra P, Khattab M, et al. Childhood acute lymphoblastic leukemia in the Middle East and neighboring countries: a prospective multi-institutional international collaborative study (CALLME1) by the Middle East Childhood Cancer Alliance (MECCA). Pediatr Blood Cancer 2014;61:1403-10. [Crossref] [PubMed]
- Tawfiq SA, Yassin AK, AlGetta HA, et al. Acute myeloblastic leukemia: Important clinical and epidemiological facts from Hiwa Hospital in Sulaimaniyah, Iraq. Iraqi J Hematol 2019;8:69-73. [Crossref]
- Case M, Matheson E, Minto L, et al. Mutation of genes affecting the RAS pathway is common in childhood acute lymphoblastic leukemia. Cancer Res 2008;68:6803-9. [Crossref] [PubMed]
- North M, Shuga J, Fromowitz M, et al. Modulation of Ras signaling alters the toxicity of hydroquinone, a benzene metabolite and component of cigarette smoke. BMC Cancer 2014;14:6. [Crossref] [PubMed]
- Busby C, Hamdan M, Ariabi E. Cancer, infant mortality and birth sex-ratio in Fallujah, Iraq 2005-2009. Int J Environ Res Public Health 2010;7:2828-37. [Crossref] [PubMed]


