
Astrocyte-derived complement C3 is activated in patients with tuberous sclerosis complex and mediates immune injury: an integrated bioinformatics analysis
Highlight box
Key findings
• This study shows that complement C3, which is increased in brains and peripheral blood from patients with TSC, mediates immune injury.
What is known and what is new?
• Many studies have demonstrated that C3 is derived from astrocytes in the central nervous system.
• We found that the C3 expression was almost colocalized on GFAP+ cells in CT of patients with TSC. The TSC2 KO U87 cell line by CRISPR/Cas9 was constructed to validate that complement C3 is derived from astrocytes in a disease model of TSC.
What is the implication, and what should change now?
• Complement C3 is a potential therapeutic strategy for treating TSC given that it is a major hub of proinflammatory activity. In the future, the therapeutic effects of complement C3 can be explored by knocking it down in astrocytes in a mouse model of TSC.
Introduction
Tuberous sclerosis complex (TSC) is caused by somatic inactivation of TSC1 or TSC2, which leads to the hyperactivation of the mammalian target of rapamycin C1 (mTORC1) signaling pathway (1). TSC is a known leading cause of many neurological manifestations, including refractory epilepsy, intellectual disability, neurodevelopmental delay, autism spectrum disorder, and a range of other behavioral and psychiatric symptoms, collectively referred to as TSC-associated neuropsychiatric disorders (2-4). Cortical tubers (CT), the most common substrate for seizures, are found in around 90% of patients with TSC (1,5,6); notably, the seizures of patients with TSC are often resistant to anti-seizure medications. The neuropsychiatric symptoms of TSC are occasionally underrated and misdiagnosed because of its complexity, which can lead to intellectual delays (1). These factors significantly impair quality of life. Despite the initiation of numerous studies, the exact pathophysiological mechanisms involved in neuropsychiatric dysfunction are yet to be further investigated.
Although mTOR inhibitors have been granted approval from the Food and drug Authority (FDA) for the treatment of patients with TSC, they show limited efficacy for neurological symptoms (7,8). Given the intractable symptoms of TSC, several studies have sought to elucidate the pathogenesis of brain lesions in TSC. It is widely acknowledged that neuroinflammation in TSC-associated brain tumors may contribute to the neuronal damage (9,10). The role of neuroinflammation in TSC-associated brain tumors has received considerable attention in recent years. However, the relationship between neuroinflammation and neurological symptoms of TSC is still unclear, and no anti-inflammatory agents for the treatment of TSC have been approved.
The advent of high-throughput technologies has made it relatively a straightforward task for researchers to explore the underlying molecular mechanisms of diseases. In this study, a microarray gene expression profile from the dataset of GSE16969 was selected. Using the xCell database (https://xcell.ucsf.edu/), we identified C3 as a hub gene, which may play an essential role in regulating immune response in the CT of TSC. We then validated the presence of complement C3 in the CT and peripheral blood of patients with TSC, and explored the source of complement C3 and the effect of excessive complement C3 on neuronal cells. We present this article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-514/rc).
Methods
Downloading and processing of transcriptome datasets of CT and normal cortex (NC)
The gene expression profiles of GSE16969 and GSE62019 of CT were downloaded from the National Center for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) website (https://www.ncbi.nlm.nih.gov/geo/). The microarray data of CT from 4 patients with TSC with refractory epilepsy (GSM424819–GSM424822) and 4 NC samples from autopsy control specimens (GSM424825–GSM424828) were used to analyze the array. Data from GSE62019, including 3 patients with TSC and 3 normal controls, were used for validation. Before being analyzed, principal component analysis (PCA) was performed to evaluate the data.
Screening for differential gene expression
Screening for differentially expressed genes (DEGs) in the samples with CT and NC was performed with the “limma” package in R software (R version 4.0.3; The R Foundation for statistical Computing, Vienna, Austria), and the DEGs identified in the 2 datasets were illustrated as volcano maps. The adjusted P value <0.05 and |log2fold change (FC)| >1.0 were used as the cut-off criteria to screen DEGs.
Enrichment analysis of DEGs
Gene Ontology (GO) analysis is a useful tool to annotate genes in biological process (BP), cellular component (CC), and molecular function (MF) (11). The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a systematic database that integrates genes, compounds, and regulatory networks in known BPs to analyze gene functions and genomes at a high level (12). In this study, the R package “clusterProfiler” was used to perform the GO and KEGG pathway analyses of the DEGs with thresholds of P<0.05 and q<0.2. The online software Ingenuity Pathway Analysis (IPA; QIAGEN. Hilden, Germany) was used to predict the most significantly affected canonical pathways based on our DEGs. The top 20 canonical pathways were determined.
Protein-protein interaction (PPI) network construction and hub gene selection
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/), is a database for predicting DEG-encoded proteins and PPI (13). Subsequently, the results of STRING were analyzed and visualized using Cytoscape software. Then, 4 topological methods including maximal clique centrality (MCC), maximum neighborhood component (MNC), degree, and density of maximum neighborhood component (DMNC) in cytoHubba, a plugin of Cytoscape, were used to rank the DEGs (14). The overlapping top 20 genes, ranked using these 4 methods, were selected as hub genes.
Validation of the hub gene at the level of messenger RNA (mRNA)
The GEO dataset GSE62019 was used for further analysis to validate hub genes at the level of mRNA. Statistical significance was set at P<0.05.
Immunohistochemistry (IHC)
IHC was used to validate the translational level of hub genes. The samples were provided by Sanbo Brain Hospital of Capital Medical University (Beijing, China). Paraffin-embedded brain tissue was sectioned at 4 µm, and mounted on pre-coated glass slides (Thermo Fisher Scientific, Waltham, MA, USA). The sections were dewaxed to expose the antigen, and incubated in citrate buffer at 95–98 ℃ for 10 minutes for antigen retrieval. The sections were then washed thrice with phosphate-buffered saline (PBS; pH 7.4), followed by incubation with 0.3% hydrogen peroxide for 30 minutes at room temperature to block endogenous peroxidase and blocking buffer [5% bovine serum albumin (BSA)] for 30 minutes to block nonspecific binding. Sections were incubated overnight at 4 ℃ with primary antibodies against complement C3 (1:1,000, monoclonal rabbit, ab200999; Abcam, Cambridge, MA, USA). The next day, the slides were washed thrice with PBS, and incubated with the secondary antibody for 1 hour at room temperature. Nuclear DNA was stained with 4',6-diamidino-2-phenylindole (DAPI). The 3,3'-diaminobenzidine (DAB) substrate solution was applied to visualize the staining of the target protein.
Immunofluorescence
To detect whether the target protein was colocalized with astrocytes, immunofluorescence was performed on paraffin-embedded sections. After antigen retrieval, the sections were blocked in 0.3% hydrogen peroxide, and 5% BSA for 30 minutes at room temperature. Then, the sections were incubated with a primary antibody mix containing C3 (1:500, ab200999, Abcam) and glial fibrillary acidic protein (GFAP; 1:200, MB0345, Abmart; Berkeley Heights, NJ, USA) overnight at 4 ℃. The next day, after being washed 3 times with PBS, the sections were incubated with secondary antibody (goat anti-mouse IgG 488, Abmart, 1:500, mixed with goat anti-rabbit IgG 488, Abmart, 1:500) for 2 hours at room temperature in the dark. Then, the nuclei DNA was stained with DAPI (Sigma-Aldrich, St. Louis, MO, USA) for 5 minutes. Finally, immunofluorescence images were captured on immunofluorescence microscope (Leica, Wetzlar, Germany).
U87 cell culture
The human astrocyte cell line U87 from the American Type Culture Collection (ATCC; Manassas, VA, USA) was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco). The cultures were incubated at 37 ℃ in 5% CO2.
Generation of U87 TSC2 CRISPR knockout (KO) cell line
The TSC2 CRISPR plasmid, which included small guide RNAs (CGACCGCAGAGCGGCGTATGGGG and CAGGTTACACTTGATCGTCAAGG for TSC2), was transiently expressed in U87 cells through electroporation method for 48 hours. The cells were single-cell sorted for mCherry expression on 96-well plates. Single clones were isolated, and TSC2 KO was verified using immunoblotting.
SH-SY5Y cell culture and treatment
The human neuronal cell line SH-SY5Y (ATCC) was cultured in DMEM/F12 (1:1) (Gibco) containing 10% FBS (Gibco), and supplemented with 1% L-glutamine (Gibco). The cultures were incubated at 37 ℃ in 5% CO2. Before treatment with complement C3 (Millipore, Burlington, MA, USA) at a concentration of 10 µg/mL, the cells were seeded in 6 well plates for 24 hours.
Quantitative real-time polymerase chain reaction for detection of gene expression
The total RNA from cultured cells was isolated by using a Total RNA Kit (R6834-01, Omega; Norcross, GA, USA) according to the manufacturer’s instructions. The concentration and purity of RNA were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to determined gene expression of C3. One microgram of total RAN was reverse-transcribed into complementary DNA (cDNA) by using a Revert Aid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). Quantitative polymerase chain reaction (qPCR) was performed using a QuantiTect SYBR Green PCR kit (Thermo Fisher Scientific). The primer sequences were as follows: C3 forward: 5'-TCACCGTCAACCACAAAGCTGCTACC-3', reverse: 5'- TTTCATAGTAGGCTCGGATCTTCCA-3'; GAPDH forward: 5'-CAAAAGGGTCATCTCC-3', reverse: 5'-CCCCAGCATCAAAGGTG-3'.
Protein extraction and western blot analysis
Cells were lysed on ice in a mixture of 1× radioimmunoprecipitation assay (RIPA) buffer with freshly added protease inhibitor and phosphatase inhibitor, and centrifugated at 12,000 g for 10 minutes at 4 ℃ to remove the precipitate. Lysates were normalized by concentration based on the results of bicinchoninic acid assay (Thermo Fisher Scientific). Each sample was resolved on 10% Bis-Tris gel and subsequently transferred onto 0.45 µm polyvinylidene fluoride membranes (Millpore, Burlington, MA, USA). After blocking with 5% nonfat milk, the membranes were incubated overnight at 4 ℃ with the following primary antibodies: anti-TSC2 (D93F12; Cell Signaling Technology, Danvers, MA, USA), anti-C3 (1:1,000, monoclonal rabbit, ab200999; Abcam), anti-β-actin (TA-09; Abmart, Shanghai, China). The membranes were washed 3 times and then incubated with goat anti-mouse IgG or goat anti-rabbit IgG secondary antibodies for 1 hour. Protein expression level was visualized using the enhanced chemiluminescence kit (Solarbio, Beijing, China). Western blot results were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Enzyme-linked immunosorbent assay (ELISA) for detection of C3 in serum of patients with TSC-associated epilepsy
In this study, 11 patients with TSC-associated epilepsy were selected from Sanbo Brain Hospital of Capital Medical University (Beijing, China). Cases and controls were individually matched based on age and gender. Blood samples (4 mL) were collected in edetic acid (EDTA)-coated plastic tubes in the morning and in a fasting state, and centrifuged at 1,500 g for 10 minutes at room temperature. The supernatants were detected by using human complement C3 ELISA kit (E-EL-H6054; Elabscience, Wuhan, China) according to the manufacturer’s instructions.
Measurement of intracellular reactive oxygen species levels
Cells were pipetted into 6-well culture dishes. After treatment with complement C3, the cells were washed 3 times with a serum-free medium, incubated with 10 µmol/L dichloro-dihydro-fluorescein diacetate (DCFH-DA; S0033S; Beyotime, Beijing, China) working solution for 20 minutes, and then washed 3 times with a serum-free medium. Fluorescence signal intensity was observed using a fluorescence microscope (Leica), the percentage of positive probe was detected through flow cytometry [Becton, Dickinson, and Co. (BD) Bioscience, Franklin Lakes, NJ, USA] and analyzed using FlowJo software (BD Bioscience).
Statistical analyses
The in vitro experiments were carried out with 3 biological replicates. Data analyses were preformed using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA). Data are expressed as the mean ± standard error of the mean (SEM). For comparisons between 2 groups, the Student’s t-test was used; the correlation between C3 gene expression and cell types was examined using the Spearman’s rank correlation test. Statistical significance was set at P<0.05.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committees of Sanbo Brain Hospital, Capital Medical University (No. 2015022704) and informed consent was taken from all the patients.
Results
Evaluation of microarray and identification of DEGs
The quality of the microarray from CT and NC in the GSE16969 dataset was evaluated prior to the differential gene expression analysis (Figure S1A). In this study, a total of 455 DEGs, including 221 upregulated and 234 downregulated genes, were identified in GSE16969 (Figure S1B).
Functional enrichment analysis
To explore the potential functions of the DEGs, we performed GO and KEGG pathway analyses. GO analysis revealed that the BP terms associated with the upregulated genes were enriched in response to interferon gamma and interferon gamma-mediated signaling pathway. The CC terms associated with the upregulated genes were enriched in collagen-containing extracellular matrix. The MF terms associated with the upregulated genes were enriched in cytokine binding and immune receptor activity (Figure S2A). For the downregulated genes, BP terms were enriched in synaptic signaling transmission. The CC terms were enriched in presynapse and synaptic membrane. The MF terms were found significantly enriched in gated channel activity and ligand-gated anion channel activity (Figure S2B).
In the pathway analysis, the significantly enriched KEGG pathways of the upregulated genes were mainly related to Staphylococcus aureus infection and complement and coagulation cascades (Figure S2C), whereas the downregulated genes were involved in glutamatergic synapse and gamma-aminobutyric acid-ergic (GABAergic) synapse signaling pathways (Figure S2D). We also preformed IPA analysis with DEGs. The top 20 significantly enriched canonical pathways are displayed in Figure S2E. Of these, neuroinflammation signaling pathway and complement system were significantly activated, whereas protein kinase A signaling and endocannabinoid neuronal synapse pathway were inhibited.
PPI networks of the DEGs and hub gene selection
To further understand the interactions between the proteins encoded by the DEGs, we constructed the PPI network based on the enquiries in the online website STRING. The constructed PPI network is shown in Figure 1, whereby protein color is ranked by the logFC. The cytoHubba plugin in Cytoscape was used to identify the hub genes. After integrating the top 20 hub genes using the top 4 common methods in cytoHubba, the following 7 hub genes were selected: LPAR5, C3, C3AR1, CX3CR1, S1PR3, CCR1, and APLNR (Table 1). Of these, C3 showed the biggest difference in the CT samples compared to controls.
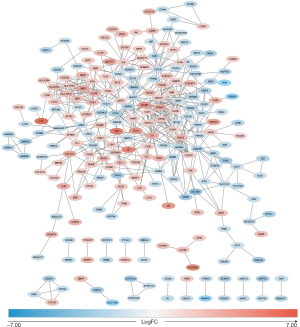
Table 1
| Rank methods in cytoHubba | Gene symbol of top 20 |
|---|---|
| MCC | GNB5, GNG12, ANXA1, LPAR5*, C3*, C3AR1*, CX3CR1*, S1PR3*, CCR1*, APLNR*, P2RY13, NPY2R, HLA-DPB1, HLA-DRA, HLA-DPA1, VCAM1, CD44, IRF8, GBP2, GBP1 |
| DMNC | P2RY13, NPY2R, APLNR*, FCGR1B, GBP3, S1PR3*, CCR1*, GBP2, GBP1, LPAR5*, C3AR1*, CHRM3, PROK2, NMB, C3*, CX3CR1*, TNC, CP, DRD1, RAMP3 |
| MNC | GNB5, GNG12, ANXA1, HLA-DPB1, HLA-DRA, HLA-DPA1, C3*, LPAR5*, CX3CR1*, VCAM1, C3AR1*, CDC42, CD44, CCR1*, S1PR3*, IRF8, P2RY13, NPY2R, APLNR*, MAPK1 |
| Degree | C3*, GNG12, CDC42, GNB5, MAPK1, ANXA1, CD44, SYK, C3AR1*, HLA-DPB1, HLA-DRA, HLA-DPA1, LPAR5*, CX3CR1*, VCAM1, IRF8, SNAP25, CCR1*, S1PR3*, APLNR* |
Genes with * symbols were the overlapped genes in the top 20 by four ranking methods respectively in cytoHubba. DEGs, differentially expressed genes; DMNC, density of maximum neighborhood component; MCC, maximal clique centrality; MNC, maximum neighborhood component.
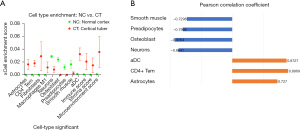
Cell-type enrichment analysis
We used the xCell database to explore correlation between the cell-type enrichment and 64 immune/stroma cell types. Our data revealed that the CT group had higher immune, stroma, and microenvironment scores than the NC group (Figure 2A). Of the 64 cell types, 9 were differentially expressed in CT compared to NC: astrocytes, CD4+ effector memory T cell (Tem), fibroblasts, macrophages M1, neurons, osteoblast, preadipocytes, smooth muscle, and activated dendritic cell (aDC); of these, 7 cell types were correlated with the expression of C3 (P<0.05) (Figure 2B).
Validation of C3 at the level of mRNA
To further validate the mRNA expression level of C3, we analyzed the DEGs in GSE62019 (samples from 3 CT and 3 NC) with R software. The relative mRNA expression of C3 in the cortical tuber samples was significant compared to NC (P<0.0001) (Figure 3).
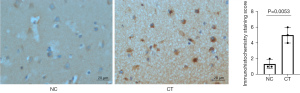
Activation of complement C3 in human TSC brain and peripheral blood
We evaluated the intensity of immunoreactive staining for complement C3 using a 0–3 scale (0= absent; 1= weak; 2= moderate; 3= strong staining) and positive area. The IHC staining of CT samples revealed a higher level of complement C3 than those of NC (Figure 4). In addition, complement C3 expression significantly increased in the peripheral blood of patients with TSC-associated epilepsy relative to that of healthy controls (Figure S3).
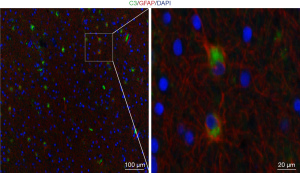
Complement C3 colocalizes on GFAP+ cells in human TSC brain
Given that the abnormal activation of the complement C3 contributes to some central nervous systems, and many studies have demonstrated that C3 derived from astrocytes, we examined C3 expression and astrocytes (GFAP+ cells). It was revealed that C3 expression was almost colocalized on GFAP+ cells (Figure 5).
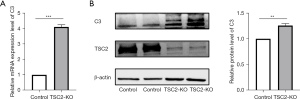
TSC2 gene deletion increased complement C3 levels in astrocytes in vitro
We established TSC2 KO U87 cells and examined C3 regulation at the RNA and protein levels. It was revealed that TSC2 deletion increased the level of C3 transcription (Figure 6A), and C3 protein expression (Figure 6B).
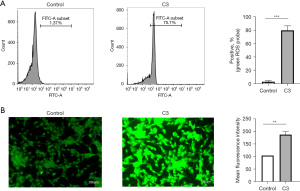
Excessive complement C3 induced immune injury in SH-SY5Y cells
To assess the immune injury induced by complement C3 on the human neuronal cell line SH-SY5Y, we measured the ROS level. Complement C3 treatment significantly increased the percentage of positive probe and the mean fluorescence intensity for ROS (Figure 7A,7B).
Discussion
The neurological complications in patients with TSC poses a great burden to their family and society because of the high morbidity and cost for treatment. Although mTOR hyperactivation is implicated in TSC neurodevelopmental features, the exact underlying molecular mechanisms and neuropathology pathways of the disorder remain poorly understood. CT are the most common lesions in the brains of patients with TSC. To gain insights into the neuropathology of TSC, we examined the molecular pathogenesis of CT using bioinformatics analysis.
We compared the transcriptome profiles of CT with those of NC and identified 455 DEGs. Based on GO and KEGG analyses, the upregulated genes in CT were mostly enriched in immune-related BPs, such as cellular response to interferon gamma, collagen-containing extracellular matrix, immune receptor activity, and complement and coagulation cascades. The downregulated genes in CT were mostly enriched in ion channel complex, glutamatergic, and GABAergic synaptic transmission. Using the IPA dataset, we found that the neuroinflammation signaling pathway and complement system were significantly activated, both of which are related to immune responses. Based on the above analysis, neuroinflammation and the complement system may present therapeutic targets for brain injury in TSC. However, the basic mechanism of disease development needs further investigation.
The finding of the correlation between immune injury and the development of CT in TSC is in accordance with reports from previous studies that identified a substantial immune system component in the development of TSC-associated brain tumors (15). The activation of inflammatory signaling pathways is one of the histopathological hallmarks of TSC (2,3). Recently, the hyperactivation of mTOR kinase has been implicated in neuroinflammation; this process likely mediates the inflammatory response in the central nervous system, and could contribute to epileptogenesis in patients with TSC (16) and other neurodevelopmental diseases (17,18). Previous studies have confirmed that proinflammatory factors are increased in the resected brain tissues of patients with TSC and TSC mouse models (15,19); for example, toll-like receptor 4 (TLR-4), interleukin 1β (IL-1β), and complement components, as well as increased expression of interleukin 17 (IL-17), tumor necrosis factor α (TNF-α), and nuclear factor κB (NF-κB) (10,20). These factors are released by the activated astrocytes and microglia cells, which represent the primary immune cells in the processes (21). Oxidative stress has been clearly observed in brains of patients with TSC and correlates with inflammation (10). An increase of ROS is believed to be a marker of oxidative stress, which can directly activate signaling of NF-κB (22). Thus, oxidative stress may be a driver of pathogenesis in the brains of patients with TSC.
We then investigated the relationship between immune cell types and the hub genes. Of the 7 identified hub genes, C3 showed the largest difference between CT and NC.
Complement C3, which is encoded by C3, is a central molecule in the innate immune pathway and plays an important role in neurobiological processes during brain development, such as elimination of synapses and neuronal migration (23). Current evidence indicates that complement C3 is implicated in neurodegenerative and neurodevelopmental diseases (24-28). Several studies have revealed that complement C3 is significantly increased in the serum of patients with autism spectrum disorder (29) and epilepsy (30), and activated in the brain of a mouse model of epilepsy (31,32), which are all TSC-associated neuropsychiatric disorders. In the context of central nervous system pathology, overactivated complement C3 contributes to neuroinflammation (33,34). Indeed, significantly increased neuroinflammation has been described in the CT of patients with TSC (10). Furthermore, several studies have confirmed that overactivation of complement C3 can induce synapse loss through complement-mediated pruning (31,35). In a recent study, loss of synapses and increased of complement component were also observed in the brains of patients with TSC (36). However, the relationship between complement-mediated synapse pruning and complement-mediated neuroinflammation has not been fully elucidated.
Several studies have demonstrated that targeting complement C3 can rescue synapse loss. In one study, C3KO mice crossed with PS2APP or TauP301S mice demonstrated reduced synapse and neuronal loss (25). In another, C3 ablation protected against synapse loss, glial activation, and reduced motor impairments while preserving cognitive function in a mouse model of experimental autoimmune encephalomyelitis (37). KO of C3 from astrocytes has been shown to significantly rescue retinal ganglion cell loss (34). Another study showed that the levels of astrocyte-derived complement C3 was increased and its presence aggravated neuronal injury (31). As mentioned, complement C3 plays a major role in the pathophysiology of several neurological disorders. Overall, the abovementioned lines of evidence support that inhibition of complement C3 may be beneficial to patients with TSC. Further studies targeting complement C3 should be conducted on a mouse model of TSC.
There were several limitations to our study. Firstly, the number of examined samples was limited because TSC is a rare disease and the collection brain samples is challenging. Second, given that transcriptome data analyses were executed in different platforms, data integration became difficult. Lastly, our analysis revealed that complement C3 is related to pro-inflammatory cell expression in the CT of TSC; however, how complement C3 mediates neuronal immune injury remains unresolved. Additional experiments are necessary to address these issues.
Conclusions
The complement system is activated in patients with TSC, and complement C3 is a critical component of the immune injury contributing to the disease.
Acknowledgments
We acknowledge GEO database for providing its platforms and contributors for uploading their meaningful datasets.
Funding: This work was supported by the Foundation of 2018 Beijing Key Clinical Specialty Construction Project-Pediatrics (No. 2199000726) and the Beijing Natural Science Foundation, China (No. S170003).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-514/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-514/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-514/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-514/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committees of Sanbo Brain Hospital, Capital Medical University (No. 2015022704) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kútna V, O'Leary VB, Newman E, et al. Revisiting Brain Tuberous Sclerosis Complex in Rat and Human: Shared Molecular and Cellular Pathology Leads to Distinct Neurophysiological and Behavioral Phenotypes. Neurotherapeutics 2021;18:845-58. [Crossref] [PubMed]
- Zimmer TS, Broekaart DWM, Gruber VE, et al. Tuberous Sclerosis Complex as Disease Model for Investigating mTOR-Related Gliopathy During Epileptogenesis. Front Neurol 2020;11:1028. [Crossref] [PubMed]
- Wong M. The role of glia in epilepsy, intellectual disability, and other neurodevelopmental disorders in tuberous sclerosis complex. J Neurodev Disord 2019;11:30. [Crossref] [PubMed]
- de Vries PJ, Wilde L, de Vries MC, et al. A clinical update on tuberous sclerosis complex-associated neuropsychiatric disorders (TAND). Am J Med Genet C Semin Med Genet 2018;178:309-20. [Crossref] [PubMed]
- van der Poest Clement E, Jansen FE, Braun KPJ, et al. Update on Drug Management of Refractory Epilepsy in Tuberous Sclerosis Complex. Paediatr Drugs 2020;22:73-84. [Crossref] [PubMed]
- Nabbout R, Belousova E, Benedik MP, et al. Epilepsy in tuberous sclerosis complex: Findings from the TOSCA Study. Epilepsia Open 2019;4:73-84. [Crossref] [PubMed]
- Kotulska K, Kwiatkowski DJ, Curatolo P, et al. Prevention of Epilepsy in Infants with Tuberous Sclerosis Complex in the EPISTOP Trial. Ann Neurol 2021;89:304-14. [Crossref] [PubMed]
- Mizuguchi M, Ikeda H, Kagitani-Shimono K, et al. Everolimus for epilepsy and autism spectrum disorder in tuberous sclerosis complex: EXIST-3 substudy in Japan. Brain Dev 2019;41:1-10. [Crossref] [PubMed]
- Zimmer TS, Ciriminna G, Arena A, et al. Chronic activation of anti-oxidant pathways and iron accumulation in epileptogenic malformations. Neuropathol Appl Neurobiol 2020;46:546-63. [Crossref] [PubMed]
- Arena A, Zimmer TS, van Scheppingen J, et al. Oxidative stress and inflammation in a spectrum of epileptogenic cortical malformations: molecular insights into their interdependence. Brain Pathol 2019;29:351-65. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Martin KR, Zhou W, Bowman MJ, et al. The genomic landscape of tuberous sclerosis complex. Nat Commun 2017;8:15816. [Crossref] [PubMed]
- Wang Z, Huang K, Yang X, et al. Downregulated GPR30 expression in the epileptogenic foci of female patients with focal cortical dysplasia type IIb and tuberous sclerosis complex is correlated with (18) F-FDG PET-CT values. Brain Pathol 2021;31:346-64. [Crossref] [PubMed]
- Abruzzo PM, Matté A, Bolotta A, et al. Plasma peroxiredoxin changes and inflammatory cytokines support the involvement of neuro-inflammation and oxidative stress in Autism Spectrum Disorder. J Transl Med 2019;17:332. [Crossref] [PubMed]
- Jiang NM, Cowan M, Moonah SN, et al. The Impact of Systemic Inflammation on Neurodevelopment. Trends Mol Med 2018;24:794-804. [Crossref] [PubMed]
- Zhang B, Zou J, Rensing NR, et al. Inflammatory mechanisms contribute to the neurological manifestations of tuberous sclerosis complex. Neurobiol Dis 2015;80:70-9. [Crossref] [PubMed]
- He JJ, Wu KF, Li S, et al. Expression of the interleukin 17 in cortical tubers of the tuberous sclerosis complex. J Neuroimmunol 2013;262:85-91. [Crossref] [PubMed]
- Vezzani A, Ravizza T, Bedner P, et al. Astrocytes in the initiation and progression of epilepsy. Nat Rev Neurol 2022;18:707-22. [Crossref] [PubMed]
- Deng W, Ding Z, Wang Y, et al. Dendrobine attenuates osteoclast differentiation through modulating ROS/NFATc1/ MMP9 pathway and prevents inflammatory bone destruction. Phytomedicine 2022;96:153838. [Crossref] [PubMed]
- Gorelik A, Sapir T, Haffner-Krausz R, et al. Developmental activities of the complement pathway in migrating neurons. Nat Commun 2017;8:15096. [Crossref] [PubMed]
- Carpanini SM, Torvell M, Morgan BP. Therapeutic Inhibition of the Complement System in Diseases of the Central Nervous System. Front Immunol 2019;10:362. [Crossref] [PubMed]
- Wu T, Dejanovic B, Gandham VD, et al. Complement C3 Is Activated in Human AD Brain and Is Required for Neurodegeneration in Mouse Models of Amyloidosis and Tauopathy. Cell Rep 2019;28:2111-2123.e6. [Crossref] [PubMed]
- Magdalon J, Mansur F, Teles E, Silva AL, et al. Complement System in Brain Architecture and Neurodevelopmental Disorders. Front Neurosci 2020;14:23. [Crossref] [PubMed]
- Petrisko TJ, Gomez-Arboledas A, Tenner AJ. Complement as a powerful "influencer" in the brain during development, adulthood and neurological disorders. Adv Immunol 2021;152:157-222. [Crossref] [PubMed]
- Rossini L, De Santis D, Cecchini E, et al. Dendritic spine loss in epileptogenic Type II focal cortical dysplasia: Role of enhanced classical complement pathway activation. Brain Pathol 2023;33:e13141. [Crossref] [PubMed]
- Shen L, Zhang K, Feng C, et al. iTRAQ-Based Proteomic Analysis Reveals Protein Profile in Plasma from Children with Autism. Proteomics Clin Appl 2018;12:e1700085. [Crossref] [PubMed]
- Kopczynska M, Zelek WM, Vespa S, et al. Complement system biomarkers in epilepsy. Seizure 2018;60:1-7. [Crossref] [PubMed]
- Jiang GT, Shao L, Kong S, et al. Complement C3 Aggravates Post-epileptic Neuronal Injury Via Activation of TRPV1. Neurosci Bull 2021;37:1427-40. [Crossref] [PubMed]
- Schartz ND, Wyatt-Johnson SK, Price LR, et al. Status epilepticus triggers long-lasting activation of complement C1q-C3 signaling in the hippocampus that correlates with seizure frequency in experimental epilepsy. Neurobiol Dis 2018;109:163-73. [Crossref] [PubMed]
- Nitkiewicz J, Borjabad A, Morgello S, et al. HIV induces expression of complement component C3 in astrocytes by NF-kappaB-dependent activation of interleukin-6 synthesis. J Neuroinflammation 2017;14:23. [Crossref] [PubMed]
- Gharagozloo M, Smith MD, Jin J, et al. Complement component 3 from astrocytes mediates retinal ganglion cell loss during neuroinflammation. Acta Neuropathol 2021;142:899-915. [Crossref] [PubMed]
- Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell 2007;131:1164-78. [Crossref] [PubMed]
- Gruber VE, Luinenburg MJ, Colleselli K, et al. Increased expression of complement components in tuberous sclerosis complex and focal cortical dysplasia type 2B brain lesions. Epilepsia 2022;63:364-74. [Crossref] [PubMed]
- Hammond JW, Bellizzi MJ, Ware C, et al. Complement-dependent synapse loss and microgliosis in a mouse model of multiple sclerosis. Brain Behav Immun 2020;87:739-50. [Crossref] [PubMed]



