
Analysis of the CHD7 gene mutations in patients of congenital heart disease with extracardiac malformations
Highlight box
Key findings
• Four novel heterozygous loss-of-function CHD7 mutations were found in four congenital heart disease (CHD) families with extracardiac malformations and 23 rare mutations were found in 1,155 sporadic CHD patients.
What is known and what is new?
• CHD7 is a transcriptional regulator that plays an important role in the development of the central nervous system, the inner ear, and the pharyngeal arches of the neural crest.
• De novo loss-of-function variants of the CHD7 gene are the genetic cause of familial CHD with extracardiac malformations and the spectrum of pathogenic CHD7 variants in sporadic CHD is expanded.
What is the implication, and what should change now?
• CHD7 variants are associated with increased risk of CHD and extracardiac malformations development and may be served as an important susceptible genetic event for CHD.
Introduction
Congenital heart disease (CHD) is the most common type of birth defect, with an incidence rate of 8.98‰ live births in China (1). The phenotypes of CHD patients are highly variable, ranging from single, localized structural defects, such as ventricular septum defect and atrial septum defect, to complex structural malformations, like tetralogy of Fallot and transposition of the great arteries. Moreover, complex congenital heart malformations are frequently accompanied with congenital extracardiac malformations, which are present in approximately 13% of newborns with CHD, including 2% of cases that are associated with a genetic syndrome such as neurodevelopmental disabilities and CHARGE (coloboma, heart defects, choanal atresia, growth retardation, genital abnormalities, and ear abnormalities) syndrome with the identification code of 214800 on Online Mendelian Inheritance in Man (OMIM) (2). These complex malformations are associated with severe CHD subtypes, and are the leading causes of death among newborns, accounting for approximately 10% of cases of fetal demise (3,4). Uncovering the cause of CHD may have a meaningful impact on the clinical outcomes and prognosis of patients.
The causes of CHD development are complex, involving environmental and genetic factors (5). Based on recurrent risk and familial inheritance, strong evidence indicates the genetic etiology of CHD (6). With advances in next-generation sequencing (NGS), the number of research on gene variants and CHD is rapidly increasing. De novo monogenic variants are identified in approximately 10% of CHD cases, and rare inherited variants and copy number variants account for approximately 1% and 25% of affected cases, respectively (7). More than 400 genes have been reported to be associated with CHD. These known genes encode heart development-related transcription factors (TF), structural proteins, cell-signaling molecules, chromatin modifiers, cilia-related proteins (8), suggesting the genetic mechanisms of CHD are complex (7). Hence, CHD patients with the same variants may exhibit different phenotypes, or even showing complete non-penetrance (no disease) (8). NKX2-5 is a TF regulating cardiomyocyte differentiation and proliferation (9,10). Damaging mutations in NKX2-5 are associated with a wide spectrum of CHD (11), and the phenotype and penetrance of these mutations may be dependent on the genetic background of the individual or interaction with other proteins (12). GATA4 and TBX5 are another TF of T-box family. Deleterious variations in TBX5 are known to cause isolated septal defects or other types of CHD in the setting of Holt-Oram syndrome (13,14).
CHD7 (chromodomain helicase DNA-binding protein 7) is a transcriptional regulator that plays an important role in the development of the central nervous system, the inner ear, and the pharyngeal arches of the neural crest (15). Mutations in this gene have been shown to be the main pathogenic cause of CHARGE syndrome (16). Approximately 85% of CHARGE syndrome patients exhibit CHD phenotypes, such as tetralogy of Fallot, double outlet of right ventricle, and atrioventricular septal defect (17). In this study, whole exome sequencing (WES) was performed in four unrelated CHD families with extracardiac malformations, and four novel de novo deleterious variants of the CHD7 gene were found, which may be the genetic cause of the development of familial CHD with extracardiac malformations. These findings expand the spectrum of pathogenic CHD7 variants. We present this article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-634/rc).
Methods
Study participants and clinical evaluation
This study was approved by the ethics committee of Fudan University Children’s Hospital (No. 2016121), and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the guardians of the affected individuals. Four unrelated CHD families with extracardiac malformations and 1,155 patients with sporadic CHD were recruited from the Children’s Hospital of Fudan University. The CHD patients and their siblings were subjected to a series of clinical evaluations, and their blood samples were collected for trio-WES, NGS, and Sanger sequencing. Reference control data of 9,197 healthy individuals from East Asian populations were collected from GnomAD v2 (Genome Aggregation Database, https://gnomad.broad institute.org/).
Whole exome sequencing
Genomic DNA samples were extracted from the peripheral venous blood of patients and their family members by using the QIAamp DNA Blood Mini Kit (Qiagen), according to the manufacturer’s protocol. WES of the genomic DNA samples was performed by Shanghai Gemple Biotech Co. Ltd., to screen for deleterious genetic variants via stringent bioinformatics analysis. Allele frequencies were obtained from GnomAD v2. Genetic variants were predicted to be deleterious by using SIFT (Sorting Intolerant from Tolerant, https://sift.bii.a-star.edu.sg/), PolyPhen2 (prediction of functional effects of human nsSNPs, http://genetics.bwh.harvard.edu/pph2/), MutationTaster (https://www.mutationtaster.org/), and CADD (Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu/snv). Unique or rare variants (absence or minor allele frequency <0.01% in gnomAD_exome_EAS) were also screen, and deleterious candidates (damaging variants as predicted by the SIFT, PolyPhen2, and MutationTaster prediction programs, and CADD score >20) were predicted.
Sanger sequencing
The selected candidate variants screened by WES were validated using Sanger sequencing in the patients and their family members. Primer pairs were designed using the online software Primer3 (https://bioinfo.ut.ee/primer3/), and were used to amplify fragments including the variants. The primers used in this study are listed in Table 1. For targeted sequencing, the sequencing of CHD7 was performed to an average depth of 100×. Mutation filtering was conducted using the same criteria used for the trio-WES analysis. Polymerase chain reaction (PCR) assays were performed using genomic DNA samples following the PrimeSTAR® Max DNA Polymerase (Takara, Shiga, Japan) standard protocol. The PCR products were sequenced by Shanghai Jie Li Biotechnology Co. Ltd. (Shanghai, China). The sequencing results were analyzed using Mutation Surveyor software (SoftGenetics, Nittany Valley, Pennsylvania, USA).
Table 1
| No. patient (family) | Position | Variant | Primers | Length (bp) |
|---|---|---|---|---|
| 1 (F1) | chr8:61693844 | c.1951_1952delAAinsT | Forward: CATTCCCCGTCGGAGCCCTTTC | 516 |
| Reverse: CACCTCATTTCATAGGCTGTAAC | ||||
| 2 (F2) | chr8:61734660 | c.2913C>G | Forward: AGGACATAGATCAAGCAAAGATC | 469 |
| Reverse: ACACACTTATATCAATCTCTGTA | ||||
| 3 (F3) | chr8:61735210 | c.3106C>T | Forward: ATATTTTGTTGAATCTAAGAG | 550 |
| Reverse: TTCATAGACACAGAAAGATAAT | ||||
| 4 (F4) | chr8:61750398 | c.4353+4_4353+12 delinsGCCCA | Forward: GCCACCTCCAAATACCATCAC | 574 |
| Reverse: CATAGGCCCCTTTTCGTAGAAG |
CHD, congenital heart disease.
Reverse transcription PCR and cDNA sequencing
Fresh blood samples were obtained from the members of the four families during their return visit. Total RNA was extracted from peripheral blood mononuclear cells in the fresh blood samples by using TRIzol reagent (Thermo Fisher Scientific, USA). Next, cDNA was synthesized from 500 ng total RNA by using PrimeScript™ RT Master Mix (Takara, China) according to the manufacturer’s protocol. With reverse transcription (RT)-PCR primer pairs (forward primer: 5'-GGCTCAGGCTAGATGTCATAGA-3' and reverse primer: 5'-TAAACGATTCCGGTCAAAACAAC-3') designed using the online software Primer3 (https://bioinfo.ut.ee/primer3/), the targeted cDNA fragments were amplified and the mRNA sequence of CHD7 were analyzed by using PCR. The PCR products were subjected to Sanger sequencing. The cDNA nucleotide sequence analysis was based on the reference sequence of CHD7 (NM_017780).
AI predictions
RoseTTAFold was accessed via the RosettaCommons web-interface server Robetta (https://robetta.bakerlab.org/submit.php) to predict the possible structure of CHD7. Subsequently, the conserved score was rated using Consurf (https://consurf.tau.ac.il/).
Statistical analysis
The association of rare deleterious CHD7 mutations with CHD was analyzed using Fisher exact test (two-sided) with GraphPad Prism 9.0 (GraphPad Software). P<0.05 was considered indicative of statistical significance.
Results
Clinical features of affected individuals in CHD families with extracardiac malformations
The clinical characteristics of the affected individuals from the four CHD families are summarized in Table 2. Patient #1 in family #1 was a 30-day-old girl who was sent to the hospital because of a heart murmur detected after birth. The patient was delivered at full term, and had a birth weight of 2,800 g. The parents of the proband were not consanguineous and manifested normal phenotypes. Echocardiography revealed atrial septal defect, patent ductus arteriosus, bicuspid aortic valve, increased left pulmonary artery flow velocity, and pulmonary hypertension. Computed tomography angiography showed coarctation of the aorta and descending right arch, aberrant left subclavian artery, atrial septal defect, patent ductus arteriosus, and pulmonary hypertension. Computed tomography results indicated left lateral ventricular enlargement. The proximal bronchus of the right upper lobe showed stenosis. Laryngoscopy suggested congenital laryngeal dysplasia. The 4th and 5th toes on the left foot were deformed (syndactyly), and the pulse of dorsal artery of the foot was unclear. A fundus examination revealed two focal masses below the right eye, approximately 1 pupillary distance apart, with clear borders; no exudation or retinal hamartoma was detected.
Table 2
| Characteristics | Patient (family) | ||||
|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | Total | |
| Sex | Female | Female | Male | Male | NA |
| Age | 30 days | 5 hours | 4 days | 10 months | NA |
| Congenital heart disease | CoA, ASD, PDA | ASD, VSD, PDA, BAV | ASD, PDA | ASD, PDA | 4/4 |
| Structural brain anomalies | Lateral ventricular enlargement (left) | Lateral ventricular enlargement (left) | Lateral ventricular enlargement (both) | 3/4 | |
| Hypothalamo-hypophyseal dysfunction and genital anomalies | Cryptorchidism, Small penis | Cryptorchidism | 2/4 | ||
| Renal /Skeletal/limb anomalies | Syndactyly | Micrognathia | 2/4 | ||
| Cranial nerve dysfunction including hearing loss | Hearing loss | Hearing loss | 2/4 | ||
| Developmental delay | + | + | 2/4 | ||
| Abnormal external, middle, or inner ears | Semicircular canal defect | 1/4 | |||
NA, not available; CoA, coarctation of aorta; ASD, atrial septal defect; PDA, patent ductus arteriosus; VSD, ventricular septal defect; BAV, bicuspid aortic valve.
Proband #2 in family #2 was a baby girl who was sent to Children’s Hospital 5 hours after birth because of congenital esophageal atresia. Her clinical data were as follows: G2P1; birth weight, 2,050 g; birth length, 45 cm; head circumference, 34 cm; gestational age, 36+6 weeks; and cesarean delivery due to intrauterine distress and hydramnios. Echocardiography revealed ventricular septal defect, atrial septal defect, patent ductus arteriosus, bicuspid aortic valve, tricuspid regurgitation, and pulmonary hypertension. Magnetic resonance imaging (MRI) showed a premature infant with occipital subdural hemorrhage, posterior horn of the left lateral ventricle hematocele, and a left lateral ventricular enlargement. The pelvis of the left kidney was slightly widened. Laryngoscopy suggested softening of the larynx. A hearing test revealed severe hearing loss in both ears.
The patient in family #3 was a 4-day-old boy who was referred to our hospital because of congenital malformations. He had been delivered at full term after his mother experienced premature rupture of membranes, and had a birth weight of 2,800 g. His parents and sister all showed normal phenotypes. Echocardiography revealed patent ductus arteriosus, atrial septal defect, and pulmonary hypertension. Brain MRI showed subdural hemorrhage, left choroid plexus hemorrhage with left lateral ventricle hematocele, left frontal and bilateral top scalp hematoma, and both-side lateral ventricular slight enlargement. B-ultrasound scans revealed that liquid occupied the space from the head of the choroidal nucleus to the head of the caudate nucleus on both sides. The middle cerebral artery resistance index increased. Brainstem auditory evoked potential testing revealed extremely severe hearing loss in the left ear and moderate hearing loss in the right ear. Laryngoscopy showed congenital hypoplasia of laryngeal cartilage. A fundus examination showed a large coloboma of choroid under both eyes. The following deformities of the external genitalia were also present: cryptorchidism and small penis.
The affected individual in family #4 was a 10-month-old boy who presented with atrial septal defect, patent ductus arteriosus, and developmental delay. His clinical manifestations were as follows: G2P2, weight 7,400 g, choking while drinking milk, not crawling, left ptosis, dysplasia of the external genitalia (cryptorchidism), normal auditory brainstem responses and otoacoustic emissions, and severe hearing loss in both ears. Computed tomography scanning of the temporal bone revealed bilateral vestibular and semicircular deformities. A plain MRI scan of the inner ear canal revealed the absence of semicircular canals. Laryngoscopy showed congenital hypoplasia of laryngeal cartilage.
Identification of de novo pathogenic variants of the CHD7 gene
To determine the potential genetic cause of CHD with extracardiac malformations in the four unrelated families, WES was performed on the four probands and their siblings or parents. Considering the normal phenotypes of the probands’ parents, we focused on de novo homozygous or compound heterozygous disease-causing variants. By using a stringent filtering strategy, we identified four novel deleterious variants of the CHD7 gene, and confirmed by Sanger sequencing that all these mutations were de novo and absent in the healthy parents and siblings of the probands. All the variants were loss-of-function mutations: the frameshift mutation c.1951_1952delAAinsT(p.L651X) in family #1, two nonsense mutations c.2913C>G (p.Y971X) in family #2 and c.3106C>T(pA1036X) in family #3, and the splicing mutation C.4353+4_4353+12delinsGCCCA in family #4 (Table 3).
Table 3
| No. patient (family) | Position | Variant | Protein change | SIFT a | PolyPhen2a | MutationTastera | CADD_phred score | ExAC_EASb | Pattern of inheritance |
|---|---|---|---|---|---|---|---|---|---|
| 1 (F1) | chr8:61693844 | c.1951_1952delAAinsT | p.L651X | NA | NA | NA | 52.3 | 0 | De novo |
| 2 (F2) | chr8:61734660 | c.2913C>G | p.Y971X | NA | NA | NA | 39.1 | 0 | De novo |
| 3 (F3) | chr8:61735210 | c.3106C>T | p.A1036X | NA | NA | NA | 41 | 0 | De novo |
| 4 (F4) | chr8:61750398 | c.4353+4_4353+12 delinsGCCCA | Splicing | NA | NA | NA | 49.6 | 0 | De novo |
a, mutation assessment by SIFT, PolyPhen-2 (PPH2), and MutationTaster. b, frequency of corresponding mutations in all populations of the ExAC Browser. NA, not available; CHD, congenital heart disease.
In family #1, the proband was found to harbor a novel heterozygous frameshift mutation in the CHD7 gene (CHD7: NM_017780, exon 3, c.1951_1952delAAinsT;p.L651X), which led to the deletion of two AA bases and the insertion of one T base, producing a stop codon and resulting in a protein that was prematurely truncated at acid amino position 651. This loss-of-function variant was absent in the public database gnomAD_genome_EAS, and was confirmed to be a de novo mutation by Sanger sequencing of the proband and his parents (Figure 1). The altered residue was located in an unknown domain of the CHD7 protein and was highly conserved across many species, except for zebrafish (Figure 2).
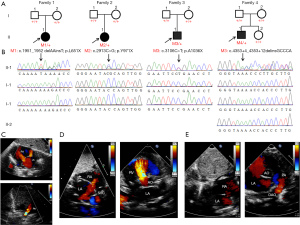
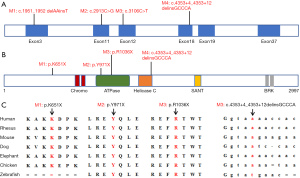
Patient #2 in family #2 was a newborn girl who carried a novel heterozygous nonsense variant in the CHD7 gene (CHD7: NM_017780, exon 11, c.2913C>G;p.Y971X). Sanger sequencing of the patient and her parents confirmed this variant to be a de novo mutation (Figure 1). The variant was located in exon 11 of the CHD7 gene and involved a stop codon, resulting in the premature truncation of the CHD7 protein at acid amino position 971 in an ATPase-related domain. This variant was predicted to be potentially deleterious. The affected residue was highly conserved among all seven species (Figure 2), supporting its pathogenicity.
In family #3, the proband was a 4-day-old boy with congenital malformations who harbored a heterozygous nonsense variant of the CHD7 gene (CHD7: NM_017780, exon 12, c.3106C>T;p.A1036X). Sanger sequencing of the proband and his parents confirmed that this variant was only present in the proband (Figure 1). The variant was located in exon 12 and produced a stop-gain change, which resulted in the premature truncation of the CHD7 protein at acid amino position 1,036 in the SNF2-related domain. The affected residue was highly conserved across many species (Figure 2). This variant was absent in the public database and was predicted to be pathogenic.
The affected individual in family #4 was a 10-month-old boy who harbored a damaging heterozygous variant affecting normal splicing (CHD7: NM_017780, exon 18, C.4353+4_4353+12delinsGCCCA). The variant was located in exon 18 and has not been reported previously. The variant was absent in the patient’s healthy parents and other family members, and was validated to be a de novo mutation by Sanger sequencing (Figure 1).
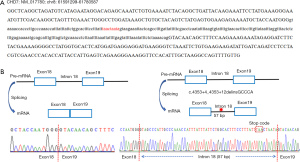
Evidence of the splice variant leading to a splicing abnormality
To verify the influence of the damaging CHD7 variant in family #4 on splicing, RT-PCR was used to synthesize cDNA from total RNA samples collected from the patient and a healthy control subject. The obtained cDNA was then used as a template and subjected to Sanger sequencing of the region spanning exons 18 and 19; finally, the results were aligned to the genomic sequence (Figure 3A). The normal control subject was found to exhibit normal precursor mRNA (pre-mRNA) splicing that only included exons 18 and 19 (Figure 3B). For the patient with the splice variant, a 57-bp fragment was retained between exons 18 and 19 in the cDNA sequence, producing a stop codon and leading to a truncated CHD7 protein (Figure 3C). These findings revealed that the damaging CHD7 variant in family #4 influenced pre-mRNA splicing, supporting the pathogenic role of this splice mutation.
Association of rare deleterious CHD7 mutations with sporadic congenital heart disease
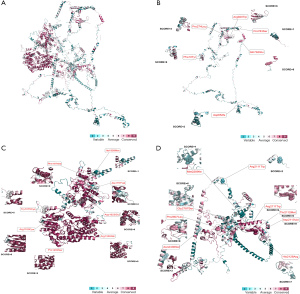
Further targeted sequencing was performed to investigate the prevalence of the disease-causing CHD7 mutations in patients with sporadic congenital heart disease. In 1,155 patients, 23 novel or rare, possible disease-causing mutations were found after filtering with the following criteria: absence on GnomAD v2 or gnomADv2_exome_EAS frequency ≤0.01% and CADD score >25. The detailed characteristics of the above mutations are listed in Table 4. These mutations were found to be equally distributed in CHD7. Through RoseTTAFold and Consurf, the conserved profile and basic structure of CHD7 were predicted (Figure 4). The majority of the rare mutations exhibited conserved profiles among different species (16 of 23 mutations were rated equal to or over 5 grades out of 10 grades).
Table 4
| No. patient | Position | Variant | Protein change | SIFT a | PolyPhen2a | MutationTastera | CADD_phred score | ExAC_EASb | Consurf score |
|---|---|---|---|---|---|---|---|---|---|
| B680 | chr8:61654020 | c.29T>A | p.Phe10Tyr | D | D | D | 25.8 | 0 | 9 |
| NO_0040 | chr8:61654731 | c.740C>T | p.Pro247Leu | D | B | D | 28.2 | 0 | 4 |
| NO_0641 | chr8:61693840 | c.1953delA | p.Asp652fs | NA | NA | NA | NA | 0 | 2 |
| B357 | chr8:61707684 | c.2236C>G | p.Gln746Glu | D | P | D | 32 | 0 | 8 |
| NO_3333;B859;NO_0068 | chr8:61713055 | c.2347C>T | p.Pro783Ser | T | B | D | 26.4 | 0.001 | 1 |
| NO_0097 | chr8:61734409 | c.2758C>T | p.Arg920Trp | D | P | D | 29.4 | 0 | 6 |
| NO_2730 | chr8:61736421 | c.3224A>G | p.Tyr1075Cys | D | D | D | 27.7 | 0.0001 | 5 |
| NO_2421 | chr8:61736495 | c.3298C>T | p.Arg1100Cys | D | D | D | 27.4 | 0 | 9 |
| NO_2344 | chr8:61742956 | c.3598C>T | p.Pro1200Ser | D | D | D | 25.4 | 0 | 9 |
| NO_0285 | chr8:61748842 | c.3989G>A | p.Arg1330Gln | D | D | D | 28.2 | 0 | 9 |
| NO_2217 | chr8:61757441 | c.4869C>A | p.Asp1623Glu | T | B | D | 37 | 0 | 9 |
| B166 | chr8:61761133 | c.5270C>T | p.Ala1757Val | T | B | D | 28.3 | 0 | 4 |
| NO_1856 | chr8:61764789 | c.5877A>G | p.Ile1959Met | T | B | D | 32 | 0 | 1 |
| NO_3057 | chr8:61765082 | c.5920T>A | p.Phe1974Ile | D | D | D | 26.4 | 0 | 9 |
| NO_2541 | chr8:61765615 | c.6331C>T | p.Arg2111Trp | D | D | D | 27 | 0 | 9 |
| NO_1023 | chr8:61765619 | c.6335C>T | p.Thr2112Met | D | D | D | 28.4 | 0 | 9 |
| B508 | chr8:61765622 | c.6338A>T | p.Asp2113Val | D | D | D | 35 | 0 | 9 |
| B506 | chr8:61765667 | c.6383A>G | p.His2128Arg | T | B | D | 31 | 0 | 7 |
| NO_3252 | chr8:61767013 | c.6867G>C | p.Met2289Ile | T | B | D | 27.6 | 0 | 1 |
| NO_2930 | chr8:61768742 | c.7145C>T | p.Thr2382Met | T | B | D | 28.2 | 0.0003 | 2 |
| B459 | chr8:61769146 | c.7307A>G | p.Asn2436Ser | T | P | D | 25.2 | 0 | 3 |
| NO_1561 | chr8:61777857 | c.8359G>A | p.Gly2787Ser | D | D | D | 27.4 | 0 | 6 |
| B471 | chr8:61777918 | c.8420C>T | p.Pro2807Leu | D | D | D | 27.1 | 0 | 8 |
a, mutation assessment by SIFT, PolyPhen-2 (PPH2), and MutationTaster. b, frequency of corresponding mutations in all populations of the ExAC Browser. CHD, congenital heart disease; T, tolerant; P, probably damaging; D, disease causing; B, benign; NA, not available.
To further evaluate the association of rare deleterious CHD7 mutations with CHD, we compared the prevalence of the mutations in our sporadic CHD cohort with the control group in GnomAD v2-East Asians (CHD7 mutated allele frequency: 23/2,310 in our sporadic CHD cohort vs. 90/18,394 in control group). The association was considered statistically significant (P=0.0039, Fisher exact test; Table 4).
Discussion
In this study, to understand the genetic cause of CHD accompanied by extracardiac malformations, the WES data of four unrelated CHD families with extracardiac malformations were analyzed. Although there were many potentially predicted damaging variants in the affected individuals, considering the normal phenotypes of the probands’ parents and siblings, we focused on de novo homozygous or compound heterozygous disease-causing variants. Four de novo damaging variants were identified in the the CHD7 gene, including the frameshift mutation c.1951_1952delAAinsT (p.L651X) in family #1, the nonsense mutations c.2913C>G (p.Y971X) in family #2 and c.3106C>T (pA1036X) in family #3, and the splicing mutation C.4353+4_4353+12delinsGCCCA in family #4. The above findings suggest that loss-of-function CHD7 variants may be responsible for the development of familial CHD with extracardiac malformations.
CHD7 belongs to the SNF2/RAD54 helicase family, and is a type of ATP-dependent chromatin remodeling factor. During early human embryonic development, CHD7 is highly expressed in many tissues and organs, such as the heart, neural tube, undifferentiated neuroepithelium, and stroma, and controls the formation of multipotent neural crest cells by cooperating with PBAF (18). The cardiac neural crest cells play an important role in the development of the cardiac outflow tract; these cells migrate ventrally to form the third, fourth, and sixth pharyngeal arch arteries, and give rise to the septum that divides the truncus arteriosus, a single great vessel arising from the embryonic heart, into the aorta and pulmonary trunk (19). Mutations in the CHD7 gene have been proven to be associated with multiple developmental disorders, such as CHARGE syndrome (20,21). The phenotype spectrum of CHD in CHARGE syndrome varies greatly and includes tetralogy of Fallot, double outlet of right ventricle, and atrioventricular septal defect (22). In this study, all of the four patients had different CHD phenotypes. Patient #1 in family #1 showed atrial septal defect, patent ductus arteriosus, bicuspid aortic valve, and pulmonary hypertension. Patient #2 presented with atrial septal defect and patent ductus arteriosus. Proband #3 in family #3 had ventricular septal defect, atrial septal defect, patent ductus arteriosus, two-leafed aortic valve, tricuspid regurgitation, and pulmonary hypertension. The patient in family #4 had atrial septal defect, patent ductus arteriosus, and pulmonary hypertension. We analyzed the WES data of these 4 CHD families with extracardiac malformations, and found 4 loss-of-function variants of the CHD7 gene that were potentially associated with CHD development. However, some studies have suggested that CHD7 mutations are not a major cause of congenital heart defects, even when patients are selected for extracardiac features (23,24). A series of studies showed that the incidence of CHD was higher in patients of CHARGE syndrome with CHD7 mutations (66–92%) than in those without CHD7 mutations (71%) (21,25-27). Moreover, in patients with pathogenic CHD7 variants, the incidence of isolated or other disease-related atrioventricular septal defects and patent ductus arteriosus was higher (27), which is consistent with the CHD phenotype in our study.
De novo variants have been proven to play a key role in CHD development. Their contribution has been estimated to be 8% in sporadic CHD and approximately 28% in CHD patients with extracardiac malformations (28,29). In the present study, we conducted Sanger sequencing of the probands and their healthy parents and siblings, and confirmed that all the identified variants were de novo mutations and were absent in the healthy parents and siblings. Several types of mutations have been detected in the CHD7 gene, including frameshift, nonsense, splice site, and missense mutations (30). Heterozygous mutations and loss-of-function mutations of CHD7 are found in 60–70% of patients with CHARGE syndrome (21,31). For example, in a family with CHARGE syndrome, trio-WES analysis of the proband and their parents identified the de novo pathogenic variant c.4379_4380del (p.Ile1460Argfs*15) in exon 19 of the CHD7 gene, which resulted in a premature translational stop signal (32). Wang et al. reported the de novo splice-site mutation c.3523-2A>G in the CHD7 gene in a Chinese patient with typical CHARGE syndrome; this mutation activated a cryptic splice site that resulted in 172 missing base pairs in exon 15, leading to the premature truncation of the CHD7 protein (p.V1175Afs*11) (33).
In our study, all of the four identified variants in the CHD7 gene were loss-of-function mutations and were predicted to affect the function of the CHD7 protein. CHD7 is extremely intolerant to loss-of-function mutations, as demonstrated by a pLI score of 1 on DECIPHER data, which supports of the pathogenesis of the loss-of-function CHD7 variants in our study. This could explain the association of CHD7 with sporadic CHD, suggesting its function as a susceptibility gene.
Conclusions
In summary, four de novo loss-of-function mutations in the CHD7 gene in four unrelated families with CHD and extracardiac malformations are reported in this study. These findings confirm that loss-of-function CHD7 variants are responsible for the development of familial CHD with extracardiac malformations. Furthermore, the missense mutations identified in patients with sporadic CHD expand the spectrum of CHD7 variants associated with the development of CHD.
Acknowledgments
The authors wish to thank the subjects and their families for participating in the study.
Funding: This work was supported by the National Key Research and Development Program of China (Nos. 2021YFC2701000 and 2016YFC1000500); National Natural Science Foundation of China (Nos. 82270312, 81873482, 81873483, and 81800282); Shanghai Basic Research Project of Science and Technology Innovation Action Plan (No. 20JC1418300); and CAMS (Chinese Academy of Medical Sciences) Innovation Fund for Medical Sciences (No. 2019-I2M-5-002).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-634/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-634/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-634/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-634/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Children’s Hospital of Fudan University (No. 2016121) and informed consent was obtained from all the affected individuals’ guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhao L, Chen L, Yang T, et al. Birth prevalence of congenital heart disease in China, 1980-2019: a systematic review and meta-analysis of 617 studies. Eur J Epidemiol 2020;35:631-42. [Crossref] [PubMed]
- Egbe A, Lee S, Ho D, et al. Prevalence of congenital anomalies in newborns with congenital heart disease diagnosis. Ann Pediatr Cardiol 2014;7:86-91. [Crossref] [PubMed]
- Lynn MM, Salemi JL, Kostelyna SP, et al. Lesion-Specific Congenital Heart Disease Mortality Trends in Children: 1999 to 2017. Pediatrics 2022;150:e2022056294. [Crossref] [PubMed]
- Jorgensen M, McPherson E, Zaleski C, et al. Stillbirth: the heart of the matter. Am J Med Genet A 2014;164A:691-9. [Crossref] [PubMed]
- Shabana NA, Shahid SU, Irfan U. Genetic Contribution to Congenital Heart Disease (CHD). Pediatr Cardiol 2020;41:12-23. [Crossref] [PubMed]
- Cao Y, Huang R, Kong R, et al. Prevalence and risk factors for congenital heart defects among children in the Multi-Ethnic Yunnan Region of China. Transl Pediatr 2022;11:813-24. [Crossref] [PubMed]
- Zaidi S, Brueckner M. Genetics and Genomics of Congenital Heart Disease. Circ Res 2017;120:923-40. [Crossref] [PubMed]
- Yasuhara J, Garg V. Genetics of congenital heart disease: a narrative review of recent advances and clinical implications. Transl Pediatr 2021;10:2366-86. [Crossref] [PubMed]
- Robbe ZL, Shi W, Wasson LK, et al. CHD4 is recruited by GATA4 and NKX2-5 to repress noncardiac gene programs in the developing heart. Genes Dev 2022;36:468-82. [Crossref] [PubMed]
- Canac R, Cimarosti B, Girardeau A, et al. Deciphering Transcriptional Networks during Human Cardiac Development. Cells 2022;11:3915. [Crossref] [PubMed]
- Dixit R, Narasimhan C, Balekundri VI, et al. Functional analysis of novel genetic variants of NKX2-5 associated with nonsyndromic congenital heart disease. Am J Med Genet A 2021;185:3644-63. [Crossref] [PubMed]
- Gifford CA, Ranade SS, Samarakoon R, et al. Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 2019;364:865-70. [Crossref] [PubMed]
- Granados-Riveron JT, Pope M, Bu'lock FA, et al. Combined mutation screening of NKX2-5, GATA4, and TBX5 in congenital heart disease: multiple heterozygosity and novel mutations. Congenit Heart Dis 2012;7:151-9. [Crossref] [PubMed]
- Varela D, Varela T, Conceição N, et al. Functional analysis of two novel TBX5 variants present in individuals with Holt-Oram syndrome with different clinical manifestations. Mol Genet Genomics 2021;296:809-21. [Crossref] [PubMed]
- Schulz Y, Wehner P, Opitz L, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet 2014;133:997-1009. [Crossref] [PubMed]
- Ufartes R, Grün R, Salinas G, et al. CHARGE syndrome and related disorders: a mechanistic link. Hum Mol Genet 2021;30:2215-24. [Crossref] [PubMed]
- Hsu P, Ma A, Wilson M, et al. CHARGE syndrome: a review. J Paediatr Child Health 2014;50:504-11. [Crossref] [PubMed]
- Bajpai R, Chen DA, Rada-Iglesias A, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010;463:958-62. [Crossref] [PubMed]
- Kodo K, Shibata S, Miyagawa-Tomita S, et al. Regulation of Sema3c and the Interaction between Cardiac Neural Crest and Second Heart Field during Outflow Tract Development. Sci Rep 2017;7:6771. [Crossref] [PubMed]
- Krueger LA, Morris AC. Eyes on CHARGE syndrome: Roles of CHD7 in ocular development. Front Cell Dev Biol 2022;10:994412. [Crossref] [PubMed]
- Meisner JK, Martin DM. Congenital heart defects in CHARGE: The molecular role of CHD7 and effects on cardiac phenotype and clinical outcomes. Am J Med Genet C Semin Med Genet 2020;184:81-9. [Crossref] [PubMed]
- Corsten-Janssen N, Kerstjens-Frederikse WS, du Marchie Sarvaas GJ, et al. The cardiac phenotype in patients with a CHD7 mutation. Circ Cardiovasc Genet 2013;6:248-54. [Crossref] [PubMed]
- Pauli S, von Velsen N, Burfeind P, et al. CHD7 mutations causing CHARGE syndrome are predominantly of paternal origin. Clin Genet 2012;81:234-9. [Crossref] [PubMed]
- Qi Q, Yi L, Yang C, et al. Mutation analysis of the CHD7 gene in patients with congenital heart disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2008;25:637-41. [PubMed]
- Jongmans MC, Admiraal RJ, van der Donk KP, et al. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet 2006;43:306-14. [Crossref] [PubMed]
- Jyonouchi S, McDonald-McGinn DM, Bale S, et al. CHARGE (coloboma, heart defect, atresia choanae, retarded growth and development, genital hypoplasia, ear anomalies/deafness) syndrome and chromosome 22q11.2 deletion syndrome: a comparison of immunologic and nonimmunologic phenotypic features. Pediatrics 2009;123:e871-7. [Crossref] [PubMed]
- Lalani SR, Safiullah AM, Fernbach SD, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet 2006;78:303-14. [Crossref] [PubMed]
- Sevim Bayrak C, Zhang P, Tristani-Firouzi M, et al. De novo variants in exomes of congenital heart disease patients identify risk genes and pathways. Genome Med 2020;12:9. [Crossref] [PubMed]
- Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015;350:1262-6. [Crossref] [PubMed]
- Janssen N, Bergman JE, Swertz MA, et al. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat 2012;33:1149-60. [Crossref] [PubMed]
- Zentner GE, Layman WS, Martin DM, et al. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A 2010;152A:674-86. [Crossref] [PubMed]
- Gug C, Gorduza EV, Lăcătuşu A, et al. CHARGE syndrome associated with de novo (I1460Rfs(*)15) frameshift mutation of CHD7 gene in a patient with arteria lusoria and horseshoe kidney. Exp Ther Med 2020;20:479-85. [Crossref] [PubMed]
- Wang S, Lin Y, Liang P, et al. De novo Splice Site Mutation of the CHD7 Gene in a Chinese Patient with Typical CHARGE Syndrome. ORL J Otorhinolaryngol Relat Spec 2022;84:417-24. [Crossref] [PubMed]


