
CXCR7 promotes the migration of fibroblasts derived from patients with acquired laryngotracheal stenosis by NF-κB signaling
Highlight box
Key findings
• LTS had an increased CXCR7 expression but decreased E-cadherin expression in vivo and in vitro fibroblasts.
• CXCR7 agonist could inhibit E-cadherin expression by activating NF-κB signaling pathway and thereby promotes LTS derived fibroblasts’ migration, which may contribute to the pathogenesis of fibrosis in LTS.
What is known and what is new?
• The heterogeneity of fibroblasts exists between LTS and skin hypertrophic scar in pediatric patients. Specifically, fibroblasts derived from LTS demonstrated significantly faster cell proliferation and migration rates, compared to those from skin hypertrophic scars. The expression of CXCR7 was significantly higher in the fibroblasts from LTS than in those from skin hypertrophic scar.
• CXCR7 may have an effect on the expression of E-cadherin and migration of fibroblasts of LTS. CXCR7 inhibits the expression of E-cadherin and promotes fibroblast migration by NF-κB signaling pathway.
What is the implication, and what should change now?
• LTS may have an increased CXCR7 expression, which could inhibit E-cadherin expression by NF-κB signaling pathway and thereby promotes the migration of LTS derived fibroblasts. CXCR7 are expected to become a promising target for the treatment of LTS.
Introduction
Laryngotracheal stenosis (LTS) is a life-threatening disease that causes breathing difficulties and seriously impacts the quality of life, especially in children. Minimally invasive treatments, such as balloon dilation, and surgical therapies, such as end-to-end anastomosis and reconstruction, can effectively alleviate laryngeal obstruction symptoms but may have many complications (1). The prevention and treatment of postoperative complications is a major clinical problem. The crucial pathological process of LTS is fibrosis, which includes dysregulated fibroblast proliferation, migration, myofibroblast activation and abnormal extracellular matrix (ECM) deposition (2-4). Molecular mechanisms of fibrosis are being targeted to attenuate fibrosis in laboratory and clinical trials, such as TGF-β antagonist (5). Previous articles have mainly focused on inhibiting excessive deposition of ECM and associated signaling pathways. Recently, fibroblast and fibrosis heterogeneity have been found by single-cell RNA sequencing technology (6,7). It was revealed that fibroblasts from different anatomic sites have different transcriptional profiles and cell behaviors (8,9). So, the characteristics of LTS derived fibroblasts should be researched.
In our last work, we revealed the heterogeneity of fibroblasts between LTS and skin hypertrophic scar in pediatric patients (10). We found that fibroblasts derived from LTS demonstrated significantly faster cell proliferation and migration rates. The expression of CXC chemokine receptor-7 (CXCR7) was significantly higher in the fibroblasts of LTS than skin hypertrophic scar. But lower expression of Col1a1 and α-SMA in fibroblasts of LTS were identified, which we hypothesized that may be resulted from the effect of CXCR7. The functions of CXCR7 were mostly studied in the endothelial cells. A study reported that TGF-β1 could induce CXCR7 expression in lung endothelial cells (11). Overexpression of CXCR7 was found to decrease TGF-β1-induced collagen production and the expression of α-SMA by blocking the Jag1-Notch pathway. Inhibition of CXCR7 induced persistent upregulation of the Jag1-Notch signaling pathway (12). In human umbilical vein endothelial cells, CXCR7 knockdown was found to inhibit the cell proliferation. CXCR7 overexpression inhibited α-SMA expression (13), which is consistent with our findings. In the oral fibroblasts, CXCR7 was found to stimulate cell migration (14). In cancer cells, CXCR7 inhibition decreased the migration of cancer stem cells (15). In the head and neck squamous cell carcinoma cells, CXCR7 overexpression was also found to enhance the cell migration (16). So, CXCR7 was shown to be related with cell proliferation and migration. However, the functions of CXCR7 in fibroblasts of LTS have not been reported yet.
Thus, this study aims to investigate the role of CXCR7 in the fibroblasts derived from the LTS tissues. We characterized the expression of CXCR7 by RNA sequencing of LTS and studied the effects of CXCR7 on the expression of E-cadherin and migration of fibroblasts of LTS. We found out that CXCR7 inhibits the expression of E-cadherin and promotes fibroblast migration by NF-κB signaling pathway. We present this article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-118/rc).
Methods
Cell isolation and culture
Twelve patients (5 boys and 7 girls) were diagnosed with LTS and underwent the tracheal resection and T-shaped tracheal stent implantation surgery in Shanghai Children’s Hospital (Shanghai, China) from September 2019 to October 2020. Age at surgery ranged from 36 to 61 months (median age of 45 months). LTS tissues (n=12) and normal trachea tissues on the edge of them (n=12) were obtained from these patients. The specimens were washed three times with sterile 1× PBS and then treated with a 0.25% Collagenase I solution at 37 °C. The mixtures were filtered with sterile 70-µm strainer. Fibroblasts were cultured in high sugar Dulbecco’s Modified Eagle Medium (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 15% fetal bovine serum (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin, and 100 mg/L streptomycin. Fibroblasts were incubated at 37 °C with 5% CO2 in a humidified atmosphere. Primary fibroblasts of passages 2–6 were used.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Committees of human ethics and experimental safety of Shanghai Children’s Hospital (No. 2019R065) and informed consent was taken from the patients’ guardians.
RNA sequencing
LTS and normal trachea samples were acquired. RNA quantity was measured on a NanoDrop instrument (Thermo Fisher Scientific, Waltham, MA). The cDNA library was constructed using KAPA RNA-Seq Library Preparation Kit (Illumina, San Diego, CA). The cDNA samples sequencing was performed on an Illumina HiSeq 4000 sequencing platform. Raw data were analyzed using Illumina/Solexa Pipeline (Off-Line Base Caller software). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed for differentially expressed genes using the Database of Kyoto Encyclopedia of Genes and Genomes online tools (http://www.genome.jp/kegg). Hierarchical cluster analysis was performed by GeneSight-Lite 4.1.6 (BioDiscovery, El Segundo, CA, USA). The protein-protein interaction network and co-expression of the proteins encoded by the differentially expressed genes was analyzed by the GeneMANIA Cytoscape.
Quantitative PCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA concentrations were examined using the NanoDrop (ND-8000; Thermo Fisher Scientific). RNAs were reverse-transcribed into cDNA using the Moloney murine leukemia virus reverse transcriptase (M-MLV) system (Promega Biotech, Madison, WI, USA). Quantitative PCR (qPCR) was performed with an ABI 7900HT (Applied Biosystems, Foster City, CA, USA) system using SYBR Premix (Takara, Dalian, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. The data were analyzed using the comparison Ct (2−ΔΔCt) method and expressed as the fold change relative to the respective control. The primer sequences used in this study were as follows: GAPDH: forward, 5'-AGGTCGGTGTGAACGGATTTG-3'; reverse, 5'-GGGGTCGTTGATGGCAACA-3'; Cxcr7: forward, 5'-TCTGCATCTCTTCGACTACTCA-3'; reverse, 5'-GTAGAGCAGGACGCTTTTGTT-3'. E-cadherin: forward, 5'-TCATGATGCTACCCGAGGTTTG-3'; reverse, 5'-CAGAACTAGATGCAGCCGGAGA-3'.
Immunohistochemistry and immunofluorescence
After being fixed in 10% paraformaldehyde and embedded in paraffin, the specimens were cut into 5-µm-thick sections and then stained with primary antibodies against CXCR7 (1:200; Cat:ab138509; Abcam, Cambridge, United Kingdom) and E-cadherin [1:200; Cat:3195; Cell Signaling Technology (CST), Danvers, MA, USA] at 4 °C overnight. Secondary antibodies were used at room temperature for 30 min. The percentage of positive cells was analyzed with the ImageJ software (Image J; NIH, Bethesda, MD, USA).
For immunofluorescence, after fixed by 4% paraformaldehyde for 30 min, then permeabilized by 0.1% Triton X-100 for 30 min, cells were blocked with 5% bovine serum albumin (BSA) and goat serum at room temperature for 60 min. Anti-E-cadherin (1:200; Cat:3195; CST, Danvers, MA, USA) and P65 (1:200, Novus Biologicals, Centennials, CO) antibodies were used overnight at 4 °C, followed by an Alexa Fluor 488 or 555-conjugated secondary antibody. Cells were visualized by a confocal microscope (Leica, Solms, Germany).
Western blotting
Whole-cell protein lysates were lysed with radioimmunoprecipitation (RIPA) buffer containing a protease inhibitor cocktail (Roche, Basel, Switzerland). The concentrations of protein were measured by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific). Then, 40 µg protein lysate per sample was separated by 10% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.45-µm polyvinylidene difluoride membranes using a semi-dry transfer system (Trans-Blot Turbo; Bio-Rad Laboratories, Hercules, CA, USA). The membranes were incubated overnight at 4 °C with primary antibodies and then incubated with horseradish peroxidase–conjugated secondary antibody (1:4,000; Cat:7074; CST, Danvers, MA, USA). The secondary antibody was used and visualized using an electrochemiluminescence kit (Tanon Science & Technology, Shanghai, China), and a chemiluminescence imaging system (6100; Tanon Science & Technology). The antibodies used in this study were as follows: anti-phospho-ERK (1:2,000; Cat:4370; CST, Danvers, MA, USA), anti-ERK (1:2,000; Cat:4695; CST, Danvers, MA, USA); anti-phospho-P65 (1:1,000; Cat:3033; CST, Danvers, MA, USA), anti-P65 (1:1,000; Cat:8242; CST, Danvers, MA, USA); anti-CXCR7 (1:1,000; Cat:ab138509; Abcam, Cambridge, United Kingdom); anti-E-cadherin (1:1,000; Cat:3195; CST, Danvers, MA, USA).
EdU incorporation assay
Cell proliferation was determined by a EdU (5-ethynyl-2'-deoxyuridine) Cell Proliferation Assay Kit (Invitrogen, Click-iT® EdU Imaging Kits) according to the manufacturer’s protocol. Cells were incubated with EdU for 2 h, fixed by 4% paraformaldehyde (PFA) for 10 min, and treated by a Click-iT EdU reaction cocktail for 30 min. Cell nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI, 1:1,000; Sigma-Aldrich) for 10 min. The proportion of EdU positive cells was analyzed by Zeiss 710 laser-scanning microscope (Zeiss, Thornwood, NY, USA).
Cell migration assays
Cells were cultured in six-well plates. When fibroblasts grow 90% confluence, straight scratches were created in each well. Then, fibroblasts were treated with CXCR7 agonist or DMSO. The middle gap areas were observed and measured using ImageJ software (NIH, Bethesda, MD, USA) at 0, 24 and 48 hours.
For transwell assay, cells were cultured on the top transwell membranes of 24-well plates with 8 mm pore size polycarbonate transwell culture inserts (Corning, NY). CXCR7 agonist or DMSO was added to the lower chambers. After incubation, the membranes of transwell were removed and fixed. Migrated cells on the lower surface were stained with 0.5% crystal violet for several minutes. We used ImageJ software to count and measure the number of cells that migrated to the lower surface of the membrane.
siRNA knockdown studies
To transfect small interfering RNA (siRNA) oligos, Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) was used following the manufacturer’s instructions. The fibroblasts were transfected with 100 nM CXCR7 siRNA (SR311311, SR324637, OriGene Technologies, Rockville, MD) in 6-well plates. Scramble siRNA (sc-37007) was used as control.
Statistical analysis
The western blots and immunohistochemistry results were quantified by ImageJ software. Statistical software package SPSS 20.0 (SPSS, Chicago, IL, USA) was used with the Mann-Whitney non-parametric test or the Student’s t-test. The results were presented as means ± standard deviation (SD). Significant differences were defined as P value less than 0.05. All experiments were repeated at least three times.
Results
The expressions of CXCR7 and E-cadherin in LTS in vivo
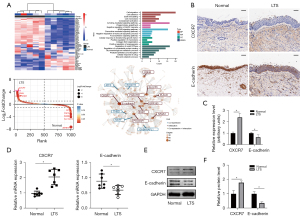
In our previous work (10), we found that the expression of CXCR7 were significantly increased in LTS. So, first, RNA sequencing was performed in acquired human LTS and normal trachea tissues to analyze differentially expressed genes and identify genes associated or co-expressed with CXCR7. Differentially expressed genes were visualized by heatmaps and gene-rank maps, which showed that CXCR7 was among the most up-regulated genes. GO and KEGG enrichment analysis found that differentially expressed genes were enriched in cell migration, cell adhesion, cytokine-mediated signaling pathway and NF-κB signaling pathway. The protein-protein interaction network and co-expression analysis showed that CXCR7 may co-expression with E-cadherin (Figure 1A). Next, we tried to investigate the expression of CXCR7 and E-cadherin in human LTS and normal trachea tissue in vivo. By immunohistochemistry, LTS showed an increased CXCR7 but decreased E-cadherin staining (Figure 1B, and the staining results were quantified in Figure 1C), which were consistent with our previous results (10) and the data of RNA sequencing. By qPCR and western blot, LTS showed an increased CXCR7 but decreased E-cadherin expression (Figure 1D-1F). These above results showed that LTS showed an increased CXCR7 expression, which may regulate the expression of E-cadherin.
CXCR7 agonist promoted cell migration of LTS derived fibroblasts in vitro
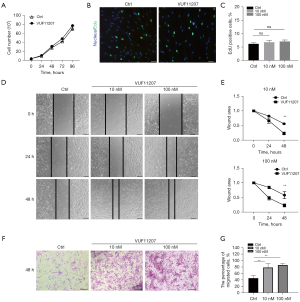
To study the functions of CXCR7 in LTS, human LTS derived fibroblasts were isolated and cultured. Fibroblasts of passages 2–6 were used. The effect of CXCR7 agonist VUF11207 on cell proliferation were analyzed by cell count and EdU incorporation assay. The results showed that CXCR7 had no effect on fibroblast proliferation (Figure 2A,2B). By PI–annexin V fluorescence-activated cell sorting analysis, there was no significant difference of the apoptotic cells after CXCR7 agonist treatment (Figure 2C). Then, wound healing assay and transwell assay were used to investigate the migration ability. The result showed that the gap area of the wound was smaller after treated by CXCR7 agonist (10 and 100 nM) for 48 hours (Figure 2D,2E). The transwell assay also found that CXCR7 agonist induced more migrated fibroblast to the lower surface of the transwell membrane (Figure 2F,2G). These above results indicated that CXCR7 agonist promoted cell migration of LTS derived fibroblasts in vitro, but not proliferation.
CXCR7 inhibited E-cadherin expression and promoted fibroblast migration through activating NF-κB signaling pathway
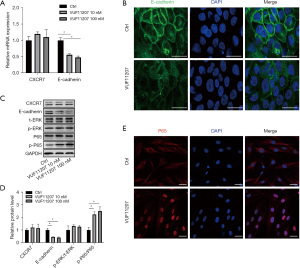
Then, we tried to explore which signaling pathway CXCR7 activated to promote fibroblast migration. It was reported that CXCR7 may enhance the migration of head and neck squamous cell carcinoma cells by upregulating TGF-β1/Smad2/3 (16) and NF-κB (17) signaling pathways. E-cadherin plays an essential role in controlling cell migration (18,19). The mRNA expressions of CXCR7 and E-cadherin were examined by qPCR. The results showed that the mRNA expressions of E-cadherin were clearly down-regulated in CXCR7 agonist treated fibroblasts, while CXCR7 expression was unchanged (Figure 3A). The decreased expression of E-cadherin in CXCR7 agonist treated fibroblasts was further confirmed by E-cadherin immunofluorescence and western blot assay (Figure 3B,3C). Next, the protein expressions of ERK and NF-κB signaling pathways were analyzed. The protein expressions of ERK and phosphorylated ERK were not significantly changed after CXCR7 agonist treatment. After CXCR7 agonist treatment (10 or 100 nM), the expression of phosphorylated P65 (a subunit of nuclear factor NF-κB) was up-regulated (Figure 3C,3D). By immunostaining of P65, P65 was shown to translocate into nuclear after CXCR7 agonist treatment, which indicated that CXCR7 activated NF-κB signaling pathway (Figure 3E).
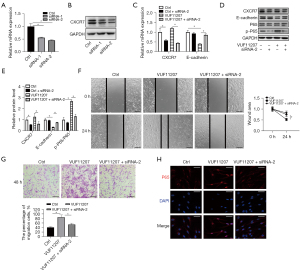
To confirm the role of CXCR7 in regulating E-cadherin expression and fibroblast migration, we inhibited CXCR7 expression by specific siRNAs and investigated the E-cadherin expression, NF-κB signaling pathway and fibroblast migration. Two siRNAs were used to inhibit the expression of CXCR7. The inhibitory effect of siRNAs was shown by qPCR and western blot assay (Figure 4A,4B). Next, we treated fibroblasts with CXCR7 siRNA and CXCR7 agonist. The results showed that CXCR7 agonist inhibited E-cadherin mRNA and protein expressions were rescued by CXCR7 siRNA significantly (Figure 4C-4E). The results of wound healing assay and transwell assays showed that CXCR7 agonist promoted fibroblast migration was inhibited by CXCR7 siRNA (Figure 4F,4G). Furthermore, NF-κB signaling pathway was examined by immunofluorescence, which showed that CXCR7 agonist activated P65 nuclear translocation decreased obviously by CXCR7 siRNA (Figure 4H). Taken together, we identified that CXCR7 inhibited E-cadherin expression by NF-κB signaling pathway and thereby promoted LTS derived fibroblast migration (Figure 5).
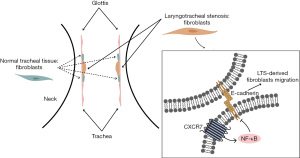
Discussion
Growing evidence showed that CXCR7 may regulate fibroblast proliferation and migration. It has been observed that CXCR7 promoted fibroblast migration in oral fibroblasts, but not proliferation (14). In lung adenocarcinoma, knockdown of CXCR7 inhibited the cell migration (15). Overexpression of CXCR7 increased human umbilical vein endothelial cells migration (13). Furthermore, CXCR7 overexpression was discovered to increase the migration ability of squamous cell carcinoma cells (16). CXCR7 blocking inhibited the migration of macrophages in the synovial tissues (20). By structural complementation assay using NanoBiT technology, SDF-1α was found to stimulate cell migration mediated by both CXCR4 and CXCR7 (21). So, CXCR7 was positively correlated with the migration of fibroblasts and other cells. Based on our previous data, we observed a decrease in the expression of E-cadherin in LTS, and we speculated that CXCR7 may regulate fibroblast migration by down-regulating E-cadherin expression (10). E-cadherin plays an essential role in controlling cell migration (18,19). In this work, we demonstrated that CXCR7 agonist stimulated the migration of LTS derived fibroblasts significantly in vitro, with no significant influence on the cell proliferation and apoptosis. Besides, we discovered that CXCR7 agonist inhibited E-cadherin expression was rescued by CXCR7 siRNA significantly. So, we confirmed the role of CXCR7 in regulating E-cadherin expression and LTS derived fibroblast migration.
In this work, we also revealed that the downstream NF-κB signaling pathway was activated by CXCR7. It has been reported that CXCR7 could activate ERK, NF-κB, Wnt/β-Catenin, TGF-β/Smad (11,15,16) or Jag1-Notch (12) signaling pathway. By using siRNA knockdown, CXCR7 was shown to activate downstream β-arrestin dependent cell signaling pathways, including ERK, p38 MAPK, and SAPK (22). In endothelial progenitor cells, shear stress stimulation increased the CXCR7 expression and induced ERK signaling pathways (23). However, in our results, ERK signaling did not significantly changed after CXCR7 agonist or siRNA knockdown treatment in the fibroblasts from LTS. In pulmonary endothelial cells, CXCR7 expression was increased and activated NF-κB signaling pathway (24). In human vascular endothelial cell line, CXCR7 inhibition blocked the phosphorylation of IKK and IκB and nuclear translocation of NF-κB P65 (25). CXCR7 enhanced the expression of tight junctions by inducing the phosphorylation of ERK and NF-κB P65 in vitro (26). In head and neck squamous cell carcinoma cells, CXCR7 overexpression was also found to enhance the migration of by upregulating TGF-β1/Smad2/3 signaling pathway (16). The authors also found that CXCR7 induced epithelial-mesenchymal transition through TGF-β1/Smad2/3 signaling pathway. However, in our previous work (10), we showed that there were no significant differences in the protein expressions of SMAD4 and phospho-SMAD2/3, suggesting that the TGF-β/Smad signaling pathway was not activated. In pulmonary capillary endothelial cells, CXCR7 overexpression decreased TGF-β1-induced collagen production and the expression of α-SMA by blocking the Jag1-Notch pathway. The inhibition of CXCR7 resulted in a persistent upregulation of the Jag1-Notch signaling pathway (12). In human umbilical vein endothelial cells in the process of angiogenesis, upregulated CXCR7 was found to inhibit the expression of α-SMA and activate AKT and ERK signaling pathways, while simultaneously suppressing Wnt/β-catenin pathways (13). In sum, CXCR7 could activate various signaling pathways such as ERK, NF-κB, Wnt/β-catenin, TGF-β/Smad or Jag1-Notch, inducing kinds of effector such as α-SMA or E-cadherin, and eventually different biological functions in specific cells or circumstances. It deserves further research to uncover the underlying factors that control the downstream signaling pathways of CXCR7 in certain cells or conditions. Nevertheless, the study has limitations relating to experimental model and translatability. The sample size of the study is relatively small. And cell culture can result in loss of context cues and alter gene expression and other attributes of in situ tissues. Additionally, there may have been confounders if the experimental tissues in the LTS and normal group were not matched optimally. What is more, the role of CXCR7-related pathways in LTS should not only be demonstrated at the cellular level, but also be verified by further animal experiments and clinical trials.
Conclusions
LTS had an increased CXCR7 expression but decreased E-cadherin expression in vivo and in vitro fibroblasts. CXCR7 agonist could inhibit E-cadherin expression by activating NF-κB signaling pathway and thereby promotes LTS derived fibroblasts’ migration, which may contribute to the pathogenesis of fibrosis in LTS.
Acknowledgments
Funding: This study was supported by National Natural Science Foundation of China (No. 82272270); and the Science and Technology Commission of Shanghai Municipality Funds (Nos. 22ZR1451700 and 21ZR1452800).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-118/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-118/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-118/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-118/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Committees of human ethics and experimental safety of Shanghai Children’s Hospital (No. 2019R065) and informed consent was taken from the patients’ guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smith MM, Buck LS. Update on the diagnosis and management of pediatric laryngotracheal stenosis. Expert Rev Respir Med 2022;16:1035-41. [Crossref] [PubMed]
- Murphy MK, Motz KM, Ding D, et al. Targeting metabolic abnormalities to reverse fibrosis in iatrogenic laryngotracheal stenosis. Laryngoscope 2018;128:E59-67. [Crossref] [PubMed]
- Hillel AT, Ding D, Samad I, et al. T-Helper 2 Lymphocyte Immunophenotype Is Associated With Iatrogenic Laryngotracheal Stenosis. Laryngoscope 2019;129:177-86. [Crossref] [PubMed]
- Motz K, Samad I, Yin LX, et al. Interferon-γ Treatment of Human Laryngotracheal Stenosis-Derived Fibroblasts. JAMA Otolaryngol Head Neck Surg 2017;143:1134-40. [Crossref] [PubMed]
- Antón-Pacheco JL, Usategui A, Martínez I, et al. TGF-β antagonist attenuates fibrosis but not luminal narrowing in experimental tracheal stenosis. Laryngoscope 2017;127:561-7. [Crossref] [PubMed]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 2018;362:eaar2971. [Crossref] [PubMed]
- Guerrero-Juarez CF, Dedhia PH, Jin S, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun 2019;10:650. [Crossref] [PubMed]
- Lynch MD, Watt FM. Fibroblast heterogeneity: implications for human disease. J Clin Invest 2018;128:26-35. [Crossref] [PubMed]
- Kotaru C, Schoonover KJ, Trudeau JB, et al. Regional fibroblast heterogeneity in the lung: implications for remodeling. Am J Respir Crit Care Med 2006;173:1208-15. [Crossref] [PubMed]
- Hu B, Wang J, Chen J, et al. The heterogeneity of fibroblasts in laryngotracheal stenosis and skin hypertrophic scar in pediatric patients. Int J Pediatr Otorhinolaryngol 2021;145:110709. [Crossref] [PubMed]
- Guan S, Zhou J. CXCR7 attenuates the TGF-β-induced endothelial-to-mesenchymal transition and pulmonary fibrosis. Mol Biosyst 2017;13:2116-24. [Crossref] [PubMed]
- Cao Z, Lis R, Ginsberg M, et al. Targeting of the pulmonary capillary vascular niche promotes lung alveolar repair and ameliorates fibrosis. Nat Med 2016;22:154-62. [Crossref] [PubMed]
- Shen M, Feng Y, Wang J, et al. CXCR7 Inhibits Fibrosis via Wnt/β-Catenin Pathways during the Process of Angiogenesis in Human Umbilical Vein Endothelial Cells. Biomed Res Int 2020;2020:1216926. [Crossref] [PubMed]
- Buskermolen JK, Roffel S, Gibbs S. Stimulation of oral fibroblast chemokine receptors identifies CCR3 and CCR4 as potential wound healing targets. J Cell Physiol 2017;232:2996-3005. [Crossref] [PubMed]
- Wu YC, Tang SJ, Sun GH, et al. CXCR7 mediates TGFβ1-promoted EMT and tumor-initiating features in lung cancer. Oncogene 2016;35:2123-32. [Crossref] [PubMed]
- Kim N, Ryu H, Kim S, et al. CXCR7 promotes migration and invasion in head and neck squamous cell carcinoma by upregulating TGF-β1/Smad2/3 signaling. Sci Rep 2019;9:18100. [Crossref] [PubMed]
- Li R, Guan Z, Bi S, et al. The proton-activated G protein-coupled receptor GPR4 regulates the development of osteoarthritis via modulating CXCL12/CXCR7 signaling. Cell Death Dis 2022;13:152. [Crossref] [PubMed]
- Grimaldi C, Schumacher I, Boquet-Pujadas A, et al. E-cadherin focuses protrusion formation at the front of migrating cells by impeding actin flow. Nat Commun 2020;11:5397. [Crossref] [PubMed]
- Cai D, Chen SC, Prasad M, et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 2014;157:1146-59. [Crossref] [PubMed]
- Kuang L, Wu J, Su N, et al. FGFR3 deficiency enhances CXCL12-dependent chemotaxis of macrophages via upregulating CXCR7 and aggravates joint destruction in mice. Ann Rheum Dis 2020;79:112-22. [Crossref] [PubMed]
- Nguyen HT, Reyes-Alcaraz A, Yong HJ, et al. CXCR7: a β-arrestin-biased receptor that potentiates cell migration and recruits β-arrestin2 exclusively through Gβγ subunits and GRK2. Cell Biosci 2020;10:134. [Crossref] [PubMed]
- Décaillot FM, Kazmi MA, Lin Y, et al. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem 2011;286:32188-97. [Crossref] [PubMed]
- Zhou H, Tu Q, Zhang Y, et al. Shear stress improves the endothelial progenitor cell function via the CXCR7/ERK pathway axis in the coronary artery disease cases. BMC Cardiovasc Disord 2020;20:403. [Crossref] [PubMed]
- Ngamsri KC, Müller A, Bösmüller H, et al. The Pivotal Role of CXCR7 in Stabilization of the Pulmonary Epithelial Barrier in Acute Pulmonary Inflammation. J Immunol 2017;198:2403-13. [Crossref] [PubMed]
- Zhao K, Yao Y, Luo X, et al. LYG-202 inhibits activation of endothelial cells and angiogenesis through CXCL12/CXCR7 pathway in breast cancer. Carcinogenesis 2018;39:588-600. [Crossref] [PubMed]
- Ngamsri KC, Jans C, Putri RA, et al. Inhibition of CXCR4 and CXCR7 Is Protective in Acute Peritoneal Inflammation. Front Immunol 2020;11:407. [Crossref] [PubMed]


