
Calcium-sensing receptor alleviates gut damage caused by endotoxemia by regulating gut microbiota
Highlight box
Key findings
• Activation of calcium-sensing receptor (CaSR) could attenuate gut injury by modulating the gut microbiome.
What is known and what is new?
• The CaSR is a central modulator of tissue response. CaSR is highly expressed in the mammalian gut.
• Activation of CaSR could enhance the species richness of the gut microbiome.
What is the implication, and what should change now?
• CaSR may impact health by supporting the establishment of a healthy gut microbiome.
Introduction
Neonatal endotoxemia remains a critical cause of mortality and morbidity in infants (1). The pathogen of this condition is primarily bacteria (2). Currently, the treatment of endotoxemia is limited to antibiotics and supportive care, and there is no specific treatment for immune dysfunction.
The main component of endotoxin is lipopolysaccharide (LPS), which is a chief component of the cell wall of gram-negative bacteria that can cause endotoxemia, induce septic shock and cause damage to multiple organs (3-5). The heart and intestines are the organs most affected by endotoxemia. Myocardial damage caused by endotoxemia is the prime reason for patient death in the intensive care unit (6). Intestinal dysfunction is one of the most common complications of endotoxemia. It can result in bacterial overgrowth and translocation and ultimately causes organ failure and even death (7).
Intestinal microecology is indeed the most intricate microecosystem in the human body. The composition of the gut microbiota is critical for its important roles in nutrition, immune regulation, and vitamin biosynthesis (8). Colonization of the gut microbiota begins during the fetal stage (9). Early in life, the gut microbiota is dynamic and influenced by environmental variables and the host (10). At approximately one year of age, the gut microecology gradually tends to be similar to that of an adult (11). There is currently no precise definition of the composition of a healthy gastrointestinal (GI) microbiota at different stages, and the key factors affecting the GI microbiota in early life are unknown (12).
Calcium-sensing receptor (CaSR) belongs to the G protein-coupled receptor (GPR) C family. It is widely expressed in various tissues and species and manipulates calcium homeostasis and osmotic balance (13,14). Our previous research demonstrated the involvement of CaSR in endotoxin myocardial injury (15). Increasing evidence notes the key role of CaSR activation in the development of various cardiovascular and renal diseases (13). CaSR is widely expressed in the digestive tract, regulates GI motility and fluid transport, participates in intestinal immunity and inflammation, and may influence the composition and balance of the intestinal microenvironment (16). A CaSR agonist (R-568) reversed the intestinal fluid secretion triggered by the cholera toxin and endotoxin of heat-resistant Escherichia coli enterotoxin by stimulating the degradation of cyclic nucleotides by phosphodiesterase, offering a novel option for treating secretory diarrhea (17). A variety of metabolites in the intestinal tract activate and affect CaSR expression. The intestinal peptide responses induced by amino acids are blocked by CaSR inhibitors and enhanced by CaSR agonists (18). We present this article in accordance with the ARRIVE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-327/rc).
Methods
Animals
Wistar rats provided by the Animal Experiment Center of Harbin Medical University (license: SYXK-2017-003) were reared in a clean-grade room under 12-h light/dark cycles (22±2 ℃) with a standard diet and water. All measures were taken to minimize the pain of the rats.
Ethical statement
Experiments were performed under a project license (IACUC approval number: 2021066) granted by the ethics board of The First Affiliated Hospital of Harbin Medical University, in compliance with institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Construction of an endotoxemia model and study design
Forty-six 2-day-old neonatal rats were randomized to the control group, LPS group, CaSR agonist group, and CaSR inhibitor group. According to the established neonatal rat model of endotoxemia, LPS (10 mg/kg) was injected intraperitoneally (19-21). An equal quantity of normal saline was given to the control rats. Rats in the agonist group were given R-568 solution (10 mg/kg, HY-10167A, MCE, Salt Lake City, USA) 1 hour after LPS injection, while rats in the inhibitor group were given Calhex-231 solution (10 mg/kg, SML0668, Sigma, St. Louis, USA) 1 hour after LPS injection. Approximately 30 minutes after the administration of LPS, systemic inflammation was assessed by observing changes in lethargy, motor activity, and shivering to confirm the successful establishment of the animal model. Rats were sacrificed by spinal dislocation after intraperitoneal injection of 10% chloral hydrate solution 24 hours or 7 days after intervention.
Intestinal sample collection
A total of 46 rat intestinal samples were obtained, including six from the 1st day of the control group (C1), five from the 1st day of the LPS group (LPS1), six from the 1st day of LPS + agonist (Agonist1), and six from the 1st day of LPS + inhibitor (Inhibitor1), including seven from the 7th day of the control group (C7), five from the 7th day of the LPS group (LPS7), six from the 7th day of the LPS + agonist (Agonist7), and five from the 7th day of the LPS + inhibitor (Inhibitor7). All samples were successfully extracted and sequenced for bioinformatics analysis. All 46 neonatal rats started breastfeeding after birth. Twenty-three rats were euthanized and sampled 1 day after intervention. At 7 days after intervention, the rest of the animals were subjected to the same procedure. Under anesthesia, colon contents were obtained with a sterile scalpel. Sampling was conducted in a sterile atmosphere. The samples were subsequently preserved in cryopreservation tubes at −80 ℃ for further investigation.
Intestinal injury evaluation
The GI tract was carefully removed after the abdominal incision, and the terminal ileum was removed approximately 5 cm immediately. The terminal 0.5 cm tissue of each sample was placed in 10% paraffin buffer for hematoxylin-eosin (H&E) staining. The degree of gut mucosal injury was based on Chiu et al.’s classification (22). Two well-experienced pathologists who did not know the grouping were responsible for the pathological examination.
DNA isolation and polymerase chain reaction (PCR) amplification
Microbial DNA was isolated from the intestinal contents of newborn rats to determine the purity of the DNA using 1% agarose gel electrophoresis. The bacterial ribosomal RNA gene V3–V4 sections were amplified, and specialized primers containing barcodes were produced. The AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) was used to extract the PCR product from a 2% agarose gel and to purify it in accordance with the manufacturer’s instructions. Majorbio Bio-Pharm Technology Co. (Shanghai, China) used an Illumina MiSeq PE300 platform (Illumina, San Diego, CA, USA) and followed industry standard procedures to pair-end sequence the purified amplicons.
Bioinformatics analysis
We applied the Majorbio Cloud Platform (https://cloud.majorbio.com/) to analyze the 16S rRNA gene sequencing data. The paired-end reads obtained by MiSeq sequencing were spliced according to the overlapping relationship (Flash, v1.2.11).
Statistical analysis
Plots of sample community compositions were compared using principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS) according to binary Jaccard distance. The linear discriminant analysis (LDA) effect size (LEfSe) (http://huttenhower.sph.harvard.edu/LEfSe) was used to detect the substantially numerous taxa (phylum to genera) of bacteria across the different groups (LDA score >2, P<0.05). The differences in α-diversity indexes were analyzed by Student’s t-test. The Adonis test was utilized for comparing PCoA values among groups. The differences based on NMDS were analyzed using the Analysis of Similarities test. The Wilcoxon rank-sum test was performed to compare species differences between the two groups, and P<0.05 was indicative of significant differences.
Results
CaSR agonist alleviated intestinal injury induced by LPS
According to the H&E staining, the intestinal morphology of the control group was basically normal; the mucosa was intact, the outline was clear, and the degree of damage was Grade 0. In the LPS1 group, the intestinal mucosa was intact, but the villi were swollen with lodging to both sides, part of the villi fell off, there was inflammatory cell infiltration in the submucosal and submucosal muscle layers, and the degree of damage was Grade 3. In the agonist group, the intestinal muscle layer was intact, the mucosa was intact, the outline was clear, and the subepithelial space at the top of the villi was widened. The villi were irregularly arranged, the submucosal and submucosal muscular layers were infiltrated with inflammatory cells, and the degree of damage was Grade 2. In the inhibitor group, the intestinal mucosa was still intact. However, the villi and propria were shed, and the exposed capillaries were dilated. The cellular component of the lamina propria was increased, there were inflammatory cells in the submucosal mucosa, and the degree of damage was Grade 4 (Figure 1A). The CaSR agonist protected against intestinal tract injury induced by LPS.
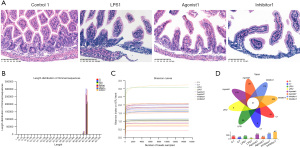
Sample sequencing data
The sequencing results revealed 2,132,963 effective sequences and 912,524,912 bp effective bases in total. The sequences had an average length of 427.84 bp. High-quality sequences with lengths between 421–440 bp accounted for 95.63% of the total sequences (Figure 1B). The alpha diversity index dilution curve plateaued with increasing data extraction volume, suggesting appropriate sequencing data volume (Figure 1C).
Species annotation and assessment
Operational taxonomic unit (OTU) analysis
In total, 1,776 OTUs were found in the groupings, which included 37 phyla, 94 classes, 220 orders, 361 families, 739 genera, and 1,202 species. Among them, there were 37 OTUs shared by each group (Figure 1D).
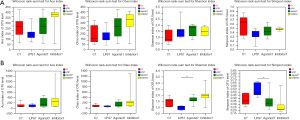
Alpha diversity analysis
The Chao and Ace indexes reflecting the abundance of the communities in the C1, Agonist1 and Inhibitor1 groups were all higher than those in the LPS1 group. In the C1 group, Agonist1 group, and Inhibitor1 group, the Simpson indexes showing community diversity were greater than those in the LPS1 group. The Shannon indexes were lower than those in the LPS1 group, but the difference was not significant (P>0.05) (Figure 2A). On the seventh day, the Shannon index of the Agonist7 group was markedly higher than that of the LPS7 group, and the Simpson index was lower than that of the LPS7 group (P<0.05). The Chao and Ace indexes of the LPS7 group were lower than those of the other three groups, but the difference was not significant (P>0.05) (Figure 2B).
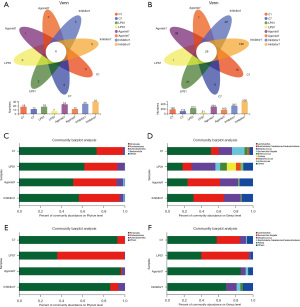
Species composition analysis
At the phylum level, there were six common phyla between the groups, namely, Bacteroidetes, Actinobacteria, Proteobacteria, Firmicutes, Chloroflexi and other unclassified phyla. The unique phylum of the Inhibitor1 group was dominated by anaerobic bacteria (Figure 3A). At the genus level, there were 29 common genera between groups, and the dominant genera were Lactobacillus and Streptococcus (Figure 3B). The unique genus of LPS7 was Succiniclasticum. The unique genera in the Agonist1 group included Halomonas, Thiobacillus, Aeribacillus and 35 other genera. At the phylum level, Firmicutes and Proteobacteria were the principal microbiota elements on the 1st day, and these were followed by Actinomycetes and Bacteroidetes. The Agonist1 group had the lowest abundance of Firmicutes and the highest abundance of Bacteroidetes (Figure 3C). At the genus level, the average Lactobacillus abundance was relatively high in the C1 group. The dominant group of the LPS1 group was Streptococcus, and Staphylococcus had the highest abundance compared with the other three groups. The dominant groups of the Agonist1 and Inhibitor1 groups were Lactobacillus, Burkholderia, and Streptococcus. The proportion of Staphylococcus in the Agonist1 group was the lowest among the four groups (Figure 3D). At the phylum level, the abundance of Firmicutes was highest in the Agonist7 group and lowest in the LPS7 group on the 7th day (Figure 3E). At the genus level, on the 7th day, the dominant bacteria of the Agonist7 group was Lactobacillus, accounting for 85.81% of bacteria. This was the highest content in the four groups, and Escherichia-Shigella was the lowest. In the LPS7 group, the content of Lactobacillus was the lowest, and the content of Escherichia-Shigella was the highest. In the Inhibitor7 group, the proportions of Rothia, Burkholderia and Streptococcus were higher than those in the other three groups (Figure 3F). The CaSR agonist increased the abundance of Lactobacillus. And it was more pronounced 7 days after the intervention.
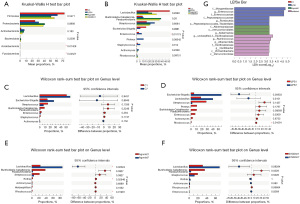
Beta diversity analysis
The difference among groups was investigated using the Adonis test. After analyzing samples taken on the 1st day, the researchers discovered greater differences in community composition across the four groups than the difference within each group (R2=0.20) (P<0.05) (Figure 4A). The Agonist1 group, Inhibitor1 group, Agonist7 group, and Inhibitor7 group were subjected to NMDS analysis using the Bray-Curtis distance algorithm, and the Adonis test was used to determine differences between groups. The results showed that the Agonist1 group and Agonist7 group (R2=0.39, P=0.001, stress =0.08) (Figure 4B) and the Inhibitor1 group and Inhibitor7 group (R2=0.25, P=0.048, stress =0.07) had significant differences (Figure 4C). CaSR affected the beta diversity of endotoxemic rats.

Species difference analysis
At the phylum level, the difference between Acidobacteriota and Fusobacteriota abundance was significant (P<0.05) (Figure 5A). At the genus level, the difference in the abundance of Enterococcus was also significant (P<0.05) (Figure 5B). At genus level, the Rhodococcus abundance in the LPS7 group was markedly lower than that in the LPS1 group. The Lactobacillus abundance in the Agonist7 group was higher than that in the Agonist1 group, and the Rhodococcus and Rothia abundances in the Agonist7 group were markedly lower than that in the Agonist1 group (P<0.05) (Figure 5C-5F). The following species were found to have statistically significant differences: the C1 group: none; the LPS1 group: Morganellaceae, Enterococcus, Enterococcaceae, and Peptostreptococcaceae; the Agonist1 group: Akkermansiaceae, Verrucomicrobiales, and Akkermansia; and the Inhibitor1 group: Oscillospiraceae, Ruminococcus, and Alloprevotella (Figure 5G). CaSR impacted the bacterial species in endotoxemic rats.
Discussion
A healthy intestinal system contains a higher abundance and more diversity of beneficial microbiota (23). The gut microbiota constitutes parts of the intestinal barrier by preventing bacterial colonization and promoting epithelial turnover (24,25). We discovered that rats with LPS injury had a high proportion of potentially pathogenic bacteria and a low proportion of helpful bacteria in the microbiota composition.
We found significantly reduced villi of the intestinal mucosa and inflammatory cell infiltration in the submucosal and submucosal muscle layers after LPS injury. Loss of epithelial barrier function is essential for the pathogenesis of inflammation and infection (26). Our findings indicated that LPS exposure destroyed the mechanical and chemical components of intestinal barriers in rats. Interestingly, we found that intestinal mucosal injury was aggravated in the inhibitor group and decreased in the agonist group.
On the 1st and 7th days, the LPS group had less diversity and richness of gut microbiota than the other groups, according to alpha diversity analysis. After LPS injury, CaSR agonists enhanced the diversity of the gut microbiota and the uniformity of the gut microbiota and helped stabilize the gut microenvironment.
The dominant phyla in the gut microbiota were Firmicutes and Proteobacteria, followed by Actinomycetes and Bacteroidetes, according to the species composition analyses. This finding is in line with prior findings (27-29). The analysis of community composition on the 1st day showed that the ratio of Firmicutes/Bacteroidetes was the lowest in the Agonist1 group. The Agonist1 group had the lowest abundance of Firmicutes and the highest abundance of Bacteroidetes. Hooper et al. documented that Bacteroidetes, prominent in normal rats and human gut microflora, regulates the levels of genes related to mucosal barrier fortification (30). Inflammatory bowel diseases, coronary heart diseases, metabolic syndromes, and other disorders are linked to an increased ratio of Firmicutes/Bacteroides, according to relevant research (31). When the ratio of Firmicutes/Bacteroides increases, the synthesis of short-chain fatty acids (SCFAs) is affected. This can result in a drop in the concentration of butyrate in the body, aggravation of the intestinal inflammatory response, and destruction of the intestinal epithelial mucosal barrier (32). The increased synthesis of SCFAs by the gut microbiota results in a decrease in pH, which decreases pathogenic species such as Escherichia coli and Enterobacteriaceae (33,34). Additionally, SCFA concentrations are sensed by specific GPRs that can mediate lipid and glucose metabolism. Furthermore, SCFAs can stimulate leukocytes, neutrophils in particular. Samuel et al. validated two orphan GPRs, GPR41 and GPR43, as receptors for SCFAs (35). SCFAs regulate the formation of dendritic cells and inflammatory factors, affecting intestinal macrophages and their immunological activity (36). The pathological findings of our study showed that activating CaSR may alleviate intestinal inflammation and maintain the integrity of the intestinal epithelial mucosa.
At the genus level, the dominant microbiota of the LPS1 group was Streptococcus, and Staphylococcus had the highest abundance compared with the other groups on the 1st day. As we know, Staphylococcus and Streptococcus can induce extensive clinical infections, from mild skin infections to deadly necrotizing fasciitis (37,38). A high abundance of Streptococcus may be one of the causes of endotoxemia susceptibility. The abundance of Streptococcus on the 1st day was markedly higher in the LPS1 group than in the other groups, and the abundance of Lactobacillus and Escherichia-Shigella increased significantly after 7 days. Therefore, it is probable that Streptococcus, Lactobacillus, and Escherichia-Shigella have a competitive interaction. In addition, we discovered that after 7 days of suckling, there was a noticeable difference in the microbiota of the LPS1 and LPS7 groups in terms of abundance and diversity. Suckling not only promotes the newborn immune response and lowers pathogen colonization, but it also promotes symbiotic colonization. This reduces intestinal injury and the danger of necrotizing enterocolitis (NEC) (37,38). Our research indicated that endotoxins reduce the colonization of Lactobacilli and induce that of pathogenic bacteria such as Streptococci and Staphylococci.
After activation of CaSR, the colonization of Lactobacillus increased, and the colonization of Rhodococcus and Rosella decreased. This difference was significant. The proportion of Staphylococcus in the Agonist1 group was the lowest among the four groups. Application of CaSR agonists reduced the colonization of intestinal pathogens. Therefore, we inferred that activating CaSR could stabilize gut microbiota injury by LPS, reduce the colonization of pathogenic bacteria, and increase the colonization of beneficial bacteria. The abundance of Lactobacilli on the 7th day of activating CaSR was significantly higher than that on the 1st day, so there may be a long-term protective effect on the gut microbiota.
After 7 days, the microbiota changed. We inferred that the abundance of Lactobacillus was the lowest in the LPS7 group, and the abundance of Escherichia-Shigella was the highest in the LPS7 group. The dominant bacterium of the Agonist7 group was Lactobacillus, accounting for 85.81% of bacteria. This was the highest content in the four groups, and the abundance of Escherichia-Shigella was the lowest. A study has shown that Lactobacillus reduces blood LPS levels by consolidating the intestinal barrier, diminishing LPS levels in feces, and repressing lipid absorption (39). Recently, epidemiologists have noticed that the high occurrence of late-onset sepsis is usually attributed to aerobic Gram-negative rods, in particular Escherichia coli, which are more likely to cause neonatal death than staphylococcal sepsis (40). In the Inhibitor7 group, the proportions of Burkholderia and Streptococcus were higher than those in the three other groups. Burkholderia consists of Gram-negative bacteria with metabolic diversity and strong adaptability. They thrive in normally hostile environments, and infections caused by these bacteria are hard to treat because they are highly resistant to antibiotics (41). This suggests that the inhibition of CaSR may lead to an increase in drug-resistant bacteria in inflammatory conditions.
The discrimination analysis by LEfSe showed that the abundance of Morganellaceae and Enterococcus in the LPS1 group had the greatest impact on the difference effect on the 1st day. Enterococcus is an essential pathogenic bacterium for nosocomial infections. Because Enterococcus has the characteristics of natural drug resistance, treatment is difficult. The application of high-dose antibiotics in early life will increase the incidence of NEC and boost the horizontal transfer of antibiotic-resistance genes (42,43). Even short-term application of antibiotics will delay the establishment of the microbiota and alter the composition of gut microbiota (12,44). Morganella bacteremia is generally related to biliary infection (45). Virtually all Morganella species strains can produce inducible chromosomal AmpC β-lactamases that hydrolyze primary and extended-spectrum penicillin and cephalosporin, causing multidrug resistance (46-48). Another finding was that in the Agonist1 group, Akkermansia had the greatest impact on the differences among the four groups. The intestinal mucus layer is a critical niche in the gut in which microbes and the host tightly interact. It is a lubricant for food to pass through the membrane, a selective barrier for nutrients to enter epithelial cells, and a protective system to prevent mechanical damage or deleterious substances (e.g., pathogens and toxins) (49). Akkermansia is a key player in the mucus-associated microbiota, and it is highly abundant in humans (50). Akkermansia is an intestinal mucin-adherent bacterium that grows by degrading mucin (51) and generates propionic acid, which is a SCFA. Additionally, Akkermansia accelerates butyrate production by promoting the growth of Anaerostipes caccae via mucin degradation (52). These SCFAs are known to promote the production of antimicrobial peptides and increase intestinal mucosal immune function (53). In addition, Akkermansia-derived extracellular vesicles that have been administered in mice are located in the large intestines and improve intestinal barrier function directly by elevating tight junction protein levels in epithelial cells (54). They are associated with healthy intestines, and the amount of Akkermansia in the gut is negatively correlated with inflammatory bowel diseases (50), appendicitis (55), obesity (56), and diabetes (57). These findings reconfirmed that CaSR activation maintains intestinal epithelial mucosal integrity by changing the gut microbiota.
Conclusions
In summary, when endotoxemia occurs, activation of CaSR may reduce the inflammatory response by diminishing the colonization of intestinal pathogens and maintaining the balance of the gut microenvironment. This discovery provides a new idea for the application of CaSR agonists and for guiding the clinical treatment and prevention of neonatal endotoxemia.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-327/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-327/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-327/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-327/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (IACUC approval number: 2021066) granted by the ethics board of The First Affiliated Hospital of Harbin Medical University, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cortese F, Scicchitano P, Gesualdo M, et al. Early and Late Infections in Newborns: Where Do We Stand? A Review. Pediatr Neonatol 2016;57:265-73. [Crossref] [PubMed]
- Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 2015;385:430-40. [Crossref] [PubMed]
- Kagan JC. Immunology. Sensing endotoxins from within. Science 2013;341:1184-5. [Crossref] [PubMed]
- Rathinam VAK, Zhao Y, Shao F. Innate immunity to intracellular LPS. Nat Immunol 2019;20:527-33. [Crossref] [PubMed]
- Lelubre C, Vincent JL. Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol 2018;14:417-27. [Crossref] [PubMed]
- Kim M, Nikouee A, Sun Y, et al. Evaluation of Parkin in the Regulation of Myocardial Mitochondria-Associated Membranes and Cardiomyopathy During Endotoxemia. Front Cell Dev Biol 2022;10:796061. [Crossref] [PubMed]
- Assimakopoulos SF, Papadopoulou I, Bantouna D, et al. Fecal Microbiota Transplantation and Hydrocortisone Ameliorate Intestinal Barrier Dysfunction and Improve Survival in a Rat Model of Cecal Ligation and Puncture-Induced Sepsis. Shock 2021;55:666-75. [Crossref] [PubMed]
- Góralczyk-Bińkowska A, Szmajda-Krygier D, Kozłowska E. The Microbiota-Gut-Brain Axis in Psychiatric Disorders. Int J Mol Sci 2022;23:11245. [Crossref] [PubMed]
- Chu DM, Ma J, Prince AL, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med 2017;23:314-26. [Crossref] [PubMed]
- Ahearn-Ford S, Berrington JE, Stewart CJ. Development of the gut microbiome in early life. Exp Physiol 2022;107:415-21. [Crossref] [PubMed]
- Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015;17:690-703. [Crossref] [PubMed]
- Chong CYL, Bloomfield FH, O'Sullivan JM. Factors Affecting Gastrointestinal Microbiome Development in Neonates. Nutrients 2018;10:274. [Crossref] [PubMed]
- Hannan FM, Kallay E, Chang W, et al. The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat Rev Endocrinol 2018;15:33-51. [Crossref] [PubMed]
- Gao Y, Robertson MJ, Rahman SN, et al. Asymmetric activation of the calcium-sensing receptor homodimer. Nature 2021;595:455-9. [Crossref] [PubMed]
- Wang HY, Liu XY, Han G, et al. LPS induces cardiomyocyte injury through calcium-sensing receptor. Mol Cell Biochem 2013;379:153-9. [Crossref] [PubMed]
- Jin T, Xu Q, Liu X, et al. Effect of Calcium-Sensitive Receptor Agonist R568 on Gastric Motility and the Underlying Mechanism. Neuroendocrinology 2023;113:289-303. [Crossref] [PubMed]
- Geibel J, Sritharan K, Geibel R, et al. Calcium-sensing receptor abrogates secretagogue- induced increases in intestinal net fluid secretion by enhancing cyclic nucleotide destruction. Proc Natl Acad Sci U S A 2006;103:9390-7. [Crossref] [PubMed]
- Mace OJ, Schindler M, Patel S. The regulation of K- and L-cell activity by GLUT2 and the calcium-sensing receptor CasR in rat small intestine. J Physiol 2012;590:2917-36. [Crossref] [PubMed]
- Fodor RŞ, Georgescu AM, Cioc AD, et al. Time- and dose-dependent severity of lung injury in a rat model of sepsis. Rom J Morphol Embryol 2015;56:1329-37.
- Guo F, Yan CY. Effect of SecinH3 on lung injury induced by sepsis of rats. Asian Pac J Trop Med 2015;8:1049-54. [Crossref] [PubMed]
- da Fonseca LM, Reboredo MM, Lucinda LM, et al. Emphysema induced by elastase enhances acute inflammatory pulmonary response to intraperitoneal LPS in rats. Int J Exp Pathol 2016;97:430-7. [Crossref] [PubMed]
- Chiu CJ, McArdle AH, Brown R, et al. Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal. Arch Surg 1970;101:478-83. [Crossref] [PubMed]
- Pickard JM, Zeng MY, Caruso R, et al. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 2017;279:70-89. [Crossref] [PubMed]
- Shi N, Li N, Duan X, et al. Interaction between the gut microbiome and mucosal immune system. Mil Med Res 2017;4:14. [Crossref] [PubMed]
- Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017;5:e1373208. [Crossref] [PubMed]
- Coleman OI, Haller D. Bacterial Signaling at the Intestinal Epithelial Interface in Inflammation and Cancer. Front Immunol 2017;8:1927. [Crossref] [PubMed]
- Sun Y, Li L, Song J, et al. Intrauterine Hypoxia Changed the Colonization of the Gut Microbiota in Newborn Rats. Front Pediatr 2021;9:675022. [Crossref] [PubMed]
- Milani C, Duranti S, Bottacini F, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev 2017;81:e00036-17. [Crossref] [PubMed]
- Del Chierico F, Vernocchi P, Petrucca A, et al. Phylogenetic and Metabolic Tracking of Gut Microbiota during Perinatal Development. PLoS One 2015;10:e0137347. [Crossref] [PubMed]
- Hooper LV, Wong MH, Thelin A, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001;291:881-4. [Crossref] [PubMed]
- Strati F, Cavalieri D, Albanese D, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017;5:24. [Crossref] [PubMed]
- Pascale A, Marchesi N, Marelli C, et al. Microbiota and metabolic diseases. Endocrine 2018;61:357-71. [Crossref] [PubMed]
- Zimmer J, Lange B, Frick JS, et al. A vegan or vegetarian diet substantially alters the human colonic faecal microbiota. Eur J Clin Nutr 2012;66:53-60. [Crossref] [PubMed]
- Glick-Bauer M, Yeh MC. The health advantage of a vegan diet: exploring the gut microbiota connection. Nutrients 2014;6:4822-38. [Crossref] [PubMed]
- Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A 2008;105:16767-72. [Crossref] [PubMed]
- Sanchez HN, Moroney JB, Gan H, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun 2020;11:60. [Crossref] [PubMed]
- Zondervan NA, Martins Dos Santos VAP, Suarez-Diez M, et al. Phenotype and multi-omics comparison of Staphylococcus and Streptococcus uncovers pathogenic traits and predicts zoonotic potential. BMC Genomics 2021;22:102. [Crossref] [PubMed]
- Tong SY, Davis JS, Eichenberger E, et al. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015;28:603-61. [Crossref] [PubMed]
- Fuke N, Nagata N, Suganuma H, et al. Regulation of Gut Microbiota and Metabolic Endotoxemia with Dietary Factors. Nutrients 2019;11:2277. [Crossref] [PubMed]
- Tan LE. Gram negative organisms and viral infections in neonatal sepsis. BMJ 2020;371:m4248. [Crossref] [PubMed]
- Lauman P, Dennis JJ. Advances in Phage Therapy: Targeting the Burkholderia cepacia Complex. Viruses 2021;13:1331. [Crossref] [PubMed]
- Zou ZH, Liu D, Li HD, et al. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann Clin Microbiol Antimicrob 2018;17:9. [Crossref] [PubMed]
- Liu J, Li Y, Feng Y, et al. Patterned progression of gut microbiota associated with necrotizing enterocolitis and late onset sepsis in preterm infants: a prospective study in a Chinese neonatal intensive care unit. PeerJ 2019;7:e7310. [Crossref] [PubMed]
- Wong WSW, Sabu P, Deopujari V, et al. Prenatal and Peripartum Exposure to Antibiotics and Cesarean Section Delivery Are Associated with Differences in Diversity and Composition of the Infant Meconium Microbiome. Microorganisms 2020;8:179. [Crossref] [PubMed]
- Song W, Sun LY, Zhu ZJ, et al. Characteristics of Gut Microbiota in Children With Biliary Atresia After Liver Transplantation. Front Physiol 2021;12:704313. [Crossref] [PubMed]
- O'Hara CM, Brenner FW, Miller JM. Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin Microbiol Rev 2000;13:534-46. [Crossref] [PubMed]
- Santiago GS, Gonçalves D, da Silva Coelho I, et al. Conjugative plasmidic AmpC detected in Escherichia coli, Proteus mirabilis and Klebsiella pneumoniae human clinical isolates from Portugal. Braz J Microbiol 2020;51:1807-12. [Crossref] [PubMed]
- Mizrahi A, Delerue T, Morel H, et al. Infections caused by naturally AmpC-producing Enterobacteriaceae: Can we use third-generation cephalosporins? A narrative review. Int J Antimicrob Agents 2020;55:105834. [Crossref] [PubMed]
- Gustafsson JK, Johansson MEV. The role of goblet cells and mucus in intestinal homeostasis. Nat Rev Gastroenterol Hepatol 2022;19:785-803. [Crossref] [PubMed]
- Rodrigues VF, Elias-Oliveira J, Pereira ÍS, et al. Akkermansia muciniphila and Gut Immune System: A Good Friendship That Attenuates Inflammatory Bowel Disease, Obesity, and Diabetes. Front Immunol 2022;13:934695. [Crossref] [PubMed]
- Ottman N, Davids M, Suarez-Diez M, et al. Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle. Appl Environ Microbiol 2017;83:e01014-17. [Crossref] [PubMed]
- Chia LW, Hornung BVH, Aalvink S, et al. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van Leeuwenhoek 2018;111:859-73. [Crossref] [PubMed]
- Tian L, Zhou XQ, Jiang WD, et al. Sodium butyrate improved intestinal immune function associated with NF-κB and p38MAPK signalling pathways in young grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol 2017;66:548-63. [Crossref] [PubMed]
- Chelakkot C, Choi Y, Kim DK, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 2018;50:e450. [Crossref] [PubMed]
- Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 2011;60:34-40. [Crossref] [PubMed]
- Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 2016;65:426-36. [Crossref] [PubMed]
- Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 2017;23:107-13. [Crossref] [PubMed]


