
RT-PCR demonstrates superior sensitivity and specificity in detecting the five neuroblastoma genes compared to the flow cytometry method for measurable residual disease
Highlight box
Key findings
• The detection of five neuroblastoma genes (NB5) can have a more accurate assessment of prognosis than flow cytometry for measurable residual disease (MRD). It represents a potential new method for better assessing disease status and guiding treatment.
What is known and what is new?
• The five NB5 genes are characterized by rare or no expression in bone marrow and peripheral blood but strong expression in neuroblastoma (NB) in vivo or in cell lines.
• Our study demonstrated that NB5 detection shows significantly higher sensitivity in assessing tumor relapse or residual disease compared to flow cytometric MRD detection. Improved NB5 assay serves as new standard to redefine disease status and is independently associated with progression-free survival in relapsed/refractory NB.
What is the implication, and what should change now?
• NB5 assay is a potential method to better guide treatment and assess prognosis. Although the number of false positives and false negatives in NB5 detection is small, larger multicenter studies that combine more clinical features and use a diversity of detection methods are still needed to comprehensively evaluate and explore the application value of these five genes in NB.
Introduction
Neuroblastoma (NB) is highly heterogeneous and has a highly variable prognosis, ranging from spontaneous regression or untreated maturation to high aggressiveness (1). Approximately 50% of pediatric patients diagnosed with NB have metastatic disease (2). The International Neuroblastoma Risk Group (INRG) reports multiple factors as independent prognostic factors for recurrence (3-5).
A variety of different therapeutic strategies are being used to improve the prognosis of patients with relapsed or refractory NB. Therapeutic targets identified using next-generation sequencing or immunology, radiation therapy, autologous stem cell transplant, targeting of the tumor microenvironment associated with promoting survival or drug resistance, and the use of the differentiating agent isotretinoin have largely improved the survival of patients with high-risk NB (6-9). However, despite intensive multimodal treatment and bone marrow (BM) transplant, the prognosis for these patients is still unsatisfactory (10). Therefore, finding more sensitive monitoring methods may be of great significance in predicting the prognosis of patients with NB.
The most common metastatic sites of NB are the BM, cortical bone, and lymph nodes. In cases of relapsed or refractory NB, most patients have BM infiltration. Although the current gold standard for BM metastasis is BM cell morphology, residual tumor cells may not be detected or clearly reflect tumor viability and metabolism as compared to progression (11,12). 123I-meta-iodobenzylguanidine (MIBG) technology can be used to assess the degree of disease progression. Unfortunately, MIBG cannot accurately quantify changes in disease sites, identify new sites of minimal disease, or distinguish parts of disease sites (13,14). Using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and droplet digital PCR (ddPCR) to evaluate the mRNA expression level of specific genes in peripheral blood (PB) and BM can provide early and rapid assessment of the response and prognosis of patients with NB at the molecular level and help to characterize areas that are not accessible by other detection methods. Related research indicates that the use of qRT-PCR or ddPCR in BM and PB at diagnosis or BM after induction treatment is capable of detecting the high expression of specific genes and of providing more comprehensive prognostic information for patients with NB (15-18).
Several studies have identified multiple individual messenger RNA (mRNA) with stable expression in NB (19-21). Due to the limited sensitivity of a single marker, a set of multiple markers was identified for the qRT-PCR testing of patients with NB (22-26). A recent study identified five genes, chromogranin A (CHGA), doublecortin (DCX), dopa decarboxylase (DDC), paired-like homeobox 2b (PHOX2B), and tyrosine hydroxylase (TH), that were rarely or never expressed in BM or PB but that were strongly expressed in NB in vivo or in cell lines (14,17). However, in previous studies, the calculation of multiple markers was primarily based on signature approach. This method of calculation can easily lead to false-negative outcomes because the expression levels of some genes do not increase. In consideration of the stability of gene expression across various risk groups of NB, their correlation with progression-free survival (PFS) (the time from patient enrollment to the onset of disease progression) or overall survival (OS) (time from enrollment to death from any cause), and their expression levels in other hematopoietic cells, the aforementioned five genes were chosen. They will be combined with appropriate calculation methods for further analysis. We detected the expression levels of five genes in different specimens [BM, PB, and cerebrospinal fluid (CSF)] and found the results were associated with clinical evaluation.
This study aimed to evaluate the sensitivity and specificity of five genes in disease progression, clarify their role in disease burden, and determine whether an NB5 assay is superior to flow cytometry of measurable residual disease (MRD) in predicting metastasis or residual disease and the prognosis of NB. Our results showed that in relapsed or refractory NB, NB5 provided earlier and more accurate detection than did flow cytometric MRD and that NB5 can be used as an independent factor for assessing NB clinical disease and prognosis. Our results can provide value in further risk stratification for more accurate assessment of NB metastasis and residual disease, better guidance of treatment, and superior assessment of prognosis. We present this article in accordance with the REMARK reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-545/rc).
Methods
Patient information
From October 2018 to June 2023, a total of 71 cases aged 1 to 13 years old (median, 5 years old) with NB were enrolled across six research centers (Hunan Provincial People’s Hospital, Children’s Hospital Affiliated to Medical College of Zhejiang University, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Anhui Provincial Children’s Hospital, Children’s Hospital of Chongqing Medical University, Children’s Hospital of Fudan University). All cases were classified according to the 2009 International Neuroblastoma Risk Group Staging System (INRGSS), the Chinese expert consensus on diagnosis and treatment of pediatric neuroblastoma (2015 or 2021 edition), or the 2007 Children’s Oncology Group (COG) Risk Staging System. They were categorized into low-risk, intermediate-risk, and high-risk groups. Among them, three patients were classified as low risk, two as intermediate risk, and the rest as high risk. All cases were diagnosed and treated according to the guidelines of the Expert Consensus on Diagnosis and Treatment of Pediatric Neuroblastoma of 2015 and the Expert Consensus on Diagnosis and Treatment of Pediatric Neuroblastoma 2021 (CCCG-NB-2021) protocol. Regular treatments were administered based on these guidelines. Three patients underwent prospective collection of BM, PB, or CSF specimens for NB5 assay. Additionally, all patients underwent concurrent BM morphology flow cytometric MRD, and qRT-PCR detection. We collected all patients with the above detect items and meeting the above criteria during the above time period for this study. A total of 182 standard disease assessments were performed and made available for analysis. The follow-up duration was 5 years, the final follow-up time was June 2023, and the median follow-up time was 14 months. During the treatment process, tumor markers were reassessed for each course of treatment, and imaging was reviewed for every two courses of treatment. Following the completion of treatment, tumor markers and imaging examinations will be reviewed every 3 months in the first year, every 4 months in the second year, and every 6 months in the third and fourth years. Follow-up was conducted by telephone interviews and outpatient review. Loss to follow-up is defined as no follow-up for more than 6 months after the end of treatment, and the last follow-up time is as of July 9, 2023. If new lesions are detected during imaging reexamination during treatment and are accompanied by an increase in neuron-specific enolase (NSE) or vanillylmandelic acid (VMA), recurrence is diagnosed. A comprehensive disease evaluation was performed on BM aspirate/biopsy prior to treatment. During the course of treatment, tumor biomarkers and BM MRD (BM-MRD) levels were examined to assess tumor burden. Additionally, the expression levels of NB5 mRNA in BM and/or PB were also tested at the same time point. The gold standard for assessing whether there is BM infiltration is BM cell morphology. This study obtained approval from the research ethics committees of each participating institution and was conducted in accordance with the approved guidelines. Investigators obtained written informed consent from the legal guardians of all the patients involved. All the aforementioned patients’ various detection parameters are compared with the qRT-PCR results to assess the correlation between qRT-PCR and other testing methods at the same time point. This analysis aims to evaluate the sensitivity, specificity, and prognostic guidance of detecting the five genes. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Medical Ethics Committee of Hunan Provincial People’s Hospital (approval No. 2023-106).
Sample processing and NB5 assay
BM, PB, or CSF total RNA extraction was performed using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) and stored at –80 ℃ until use. The RNA integrity value (RIN) was determined using a bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), with RIN >5.5 being considered acceptable. The mean RIN for BM was 8.5 (range, 6.3–9.7), and that for PB was 8.2 (range, 7.38–9.09). Reverse transcription of 2,000 ng total RNA was performed using 20 µL of reaction volume and Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase. The expression levels of CHGA, DCX, DDC, PHOX2B, and TH were quantitatively analyzed with beta-2-microglobulin (B2M) serving as the housekeeping gene for normalization. A qRT-PCR analysis was performed using predesigned and preoptimized probe and primer sets on an ABI 7500 Fast Real-time PCR system (Applied Biosystems, Thermo Fisher Scientific). Relative mRNA expression was evaluated using the Δ threshold cycle (ΔCt) method as described previously (14). The Ct value for each gene was defined as the cycle number where the amplification signal exceeded the baseline threshold by at least 0.5 cycles. If the threshold was not reached by the 45th cycle, it was designated as 45. A lower Ct value indicated a higher expression level of the NB5 genes. ddPCR was performed using a QX200 ddPCR system (BioRad Laboratories, Hercules, CA, USA) with a total volume of 20 µL. To correct for differences in total RNA quantity and complement DNA synthesis efficiency, B2M was used as a housekeeping gene to normalize the target copy numbers. Relative copy numbers of each NB5 mRNA were calculated as the copy numbers of each NB5 mRNA divided by the copy numbers of B2M mRNA multiplied by 100%. ddPCR analysis was performed following the “Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments” (27). The primer and probe sets are listed in Table S1.
Disease evaluation
BM aspirate or trephine biopsy was evaluated for the presence of NB cells during diagnosis by at least two experts. Flow cytometry was used to detect MRD. At each disease evaluation time point during BM assessment, the responses assigned included complete remission (CR) (regardless of BM infiltration at baseline, no BM infiltration was seen when BM was re-evaluated), stable disease (SD) [BM infiltration, the extent of BM infiltration is >5% on re-evaluation but does not meet the criteria for CR, very good partial remission (VGPR) and PD], partial remission (PR) (there is no infiltration in the BM, and the extent of BM infiltration is >5% during re-evaluation, or there is infiltration in the BM, and the extent of BM infiltration is >2 times and the extent of infiltration is >20% during re-evaluation), and VGPR (if any of the following conditions occurs, it will be evaluated as VGPR. The range of BM infiltration is <5%, when re-evaluated, the BM infiltrates, but it is between 0 to 5%; There is no infiltration in the BM, when re-evaluated, there is infiltration in the BM, but the range of infiltration is between 0 to 5%; BM infiltration range >20%, BM infiltration was re-evaluated but between 0 to 5%).
Statistical analysis
Statistical computations were performed using SPSS 26.0 software (IBM Corp., Armonk, NY, USA). Standard descriptive and analytic statistical methods were employed at appropriate points, including analysis of variance (ANOVA), χ2 test, and contingency table analysis (28). The significance in PFS was assessed using Kaplan-Meier survival analysis, and the two-sided log-rank test was used to compare the survival curves. Univariate analysis of predicted values of NB5 ΔCt and other variables was completed via the Cox proportional hazards regression model. Evaluate the sensitivity and specificity of NB5 detection based on the follow-up results. P values <0.05 were considered statistically significant for all tests.
Results
Patient and sample characteristics
A total of 71 newly diagnosed patients with NB who had submitted at least one specimen for NB5 assay were included in this study (Table 1). A total of 182 standard disease assessments were performed, with 113 NB5 assays performed via qRT-PCR and 24 via ddPCR. The median time between disease assessment and qRT-PCR or ddPCR detection was 2 days (Table 1, Figure S1). Among the 71 patients, there were slightly more male patients than female patients (1.63:1), the median age was 5 years old (range, 1–13 years), and 92.958% (66/71) of patients were in the high-risk group (Table 1).
Table 1
| Characteristics | Category description | Values |
|---|---|---|
| Sex | Male | 44 (61.972) |
| Female | 27 (38.028) | |
| Age (months) | – | 5 [1–13] |
| Risk group | Low-risk group | 3 (4.225) |
| Medium-risk group | 2 (2.817) | |
| High-risk group | 66 (92.958) | |
| Bone marrow morphology | Positive/total† | 8/91 (8.791) |
| MRD (flow cytometric) | Positive/total† | 15/91 (16.484) |
| Bone marrow NB5 qRT-PCR | Positive/total† | 30/89 (33.708) |
| Blood NB5 qRT-PCR | Positive/total† | 6/22 (27.273) |
| Cerebrospinal fluid NB5 qRT-PCR | Positive/total† | 2/2 (100.000) |
| Bone marrow NB5 ddPCR | Positive/total† | 13/18 (72.222) |
| Blood NB5 ddPCR | Positive/total† | 2/5 (40.000) |
| Cerebrospinal fluid NB5 ddPCR | Positive/total† | 1/1 (100.000) |
Data are shown as number (%) or median [range]. †, number of assessments. NB5, five neuroblastoma genes (CHGA, DCX, DDC, PHOX2B, and TH); CHGA, chromogranin A; DCX, doublecortin; DDC, dopa decarboxylase; PHOX2B, paired-like homeobox 2b; TH, tyrosine hydroxylase; MRD, measurable residual disease; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; ddPCR, droplet digital PCR.
Comparison of clinical biological characteristics based on the expression levels of NB5
Before conducting this study, a baseline was established using 30 negative BM specimens from the Chinese population and 100 clinical BM specimens from patients with NB. The Ct value-positive threshold was set at 39, while the ΔCt-positive threshold was set below 18. Among the five genes, a positive result was determined if there was at least one gene with a ΔCt value below 18. Patients were divided into two groups based on the expression results of NB5. A total of 38 specimens were in the NB5-positive group, and the remaining 75 specimens were in the negative group. Furthermore, we conducted a comparison of their clinical symptoms. There were no significant differences between the NB5 qRT-PCR positive group and the negative group in gender, risk group, or treatment stage. However, significant associations were found between NB5 genes and diagnosis in both the positive and negative groups (P=0.043), and there were significant differences in MRD (flow cytometry), BM morphology, and the current states of the patient (P<0.001) (Table 2).
Table 2
| Characteristics | NB5 qRT-PCR, n (%) | P value* | |
|---|---|---|---|
| Positive | Negative | ||
| Sex | 0.588 | ||
| Male | 11 (25.000) | 33 (75.000) | |
| Female | 9 (33.333) | 18 (66.667) | |
| Risk group | 0.468 | ||
| Low-risk group | 0 | 3 (100.000) | |
| Medium-risk group | 0 | 2 (100.000) | |
| High-risk group | 38 (35.185) | 70 (64.815) | |
| Treatment stage | 0.110 | ||
| Induction therapy | 30 (42.857) | 40 (57.143) | |
| Consolidation therapy | 1 (10.000) | 9 (90.000) | |
| Maintenance therapy | 7 (31.818) | 15 (68.182) | |
| End of treatment | 0 | 11 (100.000) | |
| Diagnosis | 0.043 | ||
| Primary | 22 (30.556) | 50 (69.444) | |
| Relapse | 16 (53.333) | 14 (46.667) | |
| End of treatment | 0 | 11 (100.000) | |
| MRD (flow cytometry) | <0.001 | ||
| Positive | 15 (100.000) | 0 | |
| Negative | 18 (23.684) | 58 (76.316) | |
| No disease assessments | 22 | ||
| Bone marrow morphology | <0.001 | ||
| Positive | 8 (100.000) | 0 | |
| Negative | 24 (28.916) | 59 (71.084) | |
| No disease assessments | 22 | ||
| State | <0.001 | ||
| CR | 8 (13.333) | 52 (86.667) | |
| SD | 9 (64.286) | 5 (35.714) | |
| PR | 14 (70.000) | 6 (30.000) | |
| VGPR | 7 (36.842) | 12 (63.158) | |
*, P value <0.05 indicates a statistically significant difference. MRD and bone marrow morphology assessments were not performed in 22 times each. NB5, five neuroblastoma genes (CHGA, DCX, DDC, PHOX2B, and TH); CHGA, chromogranin A; DCX, doublecortin; DDC, dopa decarboxylase; PHOX2B, paired-like homeobox 2b; TH, tyrosine hydroxylase; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; MRD, measurable residual disease; CR, complete remission; SD, stable disease; PR, partial remission; VGPR, very good partial remission.
NB5 qRT-PCR assay of both BM and PB specimens
A total of 21 paired BM and PB specimens were included in this study. It was observed that PB specimens showed 100% concordance with the BM specimens in terms of positive results. Furthermore, the BM specimens exhibited an additional five positive results compared to the PB specimens (Figure 1 and Table S2). The analysis of ΔCt values for the five genes revealed a strong correlation between BM and PB specimens. Notably, the ΔCt values in the BM were consistently smaller than those in the PB for all the genes. Each gene was positive in BM but negative in PB. These results suggest a lower concentration of NB cells in the PB than in BM. In the specimens with strong expression of the five genes by qRT-PCR (ΔCt <10), the expressions of TH, DCX, DDC, PHOX2B, and CHGA in the BM were 7.008±0.745, 5.211±1.183, 5.552±1.050, 4.309±1.256, and 4.736±1.076, respectively, which were much stronger than those in PB (Figure 1).

NB5 qRT-PCR assay of the clinical disease state in BM, PB, and CSF
To investigate the correlation between NB5 ΔCt values in different specimens and different clinical disease statuses, we performed a simple linear regression analysis. The results demonstrated a negative correlation between ΔCt values of all five genes and the tumor cell content of flow cytometric MRD (Figure 2 and Table 3). Among 88 BM specimens, 30 (34.091%) of them were positive in the NB5 assay, which was a significantly higher proportion than that of flow cytometric MRD (15/88, 17.045%). Moreover, 15 of the 30 positive NB5 assay results were consistent with the positive flow cytometric MRD results (Table S3). Additionally, there were 15 positive results detected in NB5 assay that were not detected in the flow cytometric MRD. NB5 positive results were 17.046% higher than flow cytometric MRD, and these patients were found to have different degrees of disease progression or poor treatment effect in the later stage, which was consistent with the NB5 assay.
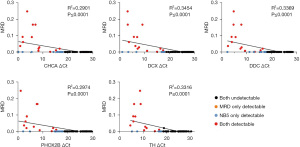
Table 3
| NB5 assay ΔCt and clinical disease | BM NB5 assay ΔCt | PB NB5 assay ΔCt | |||||
|---|---|---|---|---|---|---|---|
| Positive/total, n (%) |
Mean ΔCt for total (SE) | Mean ΔCt for positive (SE) | Positive/total, n (%) |
Mean ΔCt for total (SE) | Mean ΔCt for positive (SE) | ||
| All NB5 assays | |||||||
| CHGA | 27/88 (30.682) | 18.297 (3.073) | 11.572 (1.354) | 5/21 (23.810) | 19.239 (3.065) | 12.783 (2.084) | |
| DCX | 21/88 (23.864) | 18.387 (3.195) | 11.282 (0.602) | 4/21 (19.048) | 19.413 (2.743) | 13.747 (1.995) | |
| DDC | 25/88 (28.409) | 17.700 (2.749) | 11.673 (1.058) | 5/21 (23.810) | 19.384 (2.623) | 13.947 (1.939) | |
| PHOX2B | 23/88 (26.136) | 17.575 (3.254) | 10.366 (0.950) | 6/21 (28.571) | 20.187 (2.900) | 13.870 (1.384) | |
| TH | 19/88 (21.591) | 18.218 (3.020) | 11.591 (1.231) | 3/21 (14.286) | 20.967 (1.913) | 16.724 (0.324) | |
| Bone marrow morphology | |||||||
| CHGA | 8/88 (9.091) | 15.048 (3.235) | 7.865 (0.298) | No specimen | No specimen | No specimen | |
| DCX | 8/88 (9.091) | 16.007 (3.355) | 8.557 (0.594) | No specimen | No specimen | No specimen | |
| DDC | 8/88 (9.091) | 14.701 (3.111) | 7.855 (0.286) | No specimen | No specimen | No specimen | |
| PHOX2B | 8/88 (9.091) | 14.672 (3.525) | 6.913 (1.079) | No specimen | No specimen | No specimen | |
| TH | 8/88 (9.091) | 16.578 (2.998) | 9.971 (0.605) | No specimen | No specimen | No specimen | |
| MRD (flow cytometry) | |||||||
| CHGA | 15/88 (17.045) | 16.053 (3.326) | 8.682 (0.847) | 0/1 | – | – | |
| DCX | 15/88 (17.045) | 17.380 (3.268) | 10.200 (1.348) | 0/1 | – | – | |
| DDC | 15/88 (17.045) | 15.812 (3.103) | 9.010 (1.068) | 0/1 | – | – | |
| PHOX2B | 15/88 (17.045) | 16.099 (3.418) | 8.600 (1.436) | 1/1 (100.000) | – | – | |
| TH | 15/88 (17.045) | 17.559 (3.038) | 10.868 (1.029) | 0/1 | – | – | |
NB5, five neuroblastoma genes (CHGA, DCX, DDC, PHOX2B, and TH); ΔCt, Δ threshold cycle; BM, bone marrow; PB, peripheral blood; SE, standard error; CHGA, chromogranin A; DCX, doublecortin; DDC, dopa decarboxylase; PHOX2B, paired-like homeobox 2b; TH, tyrosine hydroxylase; MRD, measurable residual disease.
To analyze the correlation between disease burden and NB5 expression levels, the disease status of all patients was evaluated and classified as remission (CR, PR, VGPR) or SD. The expression level of NB5 in BM specimens changed with disease status. In the comparison between the remission stable groups in BM specimens, all five genes showed significant differences, with the stable group exhibiting lower ΔCt values (higher expression) compared to the remission group (Figure 3). In the comparison between the high-risk and low/medium-risk groups, only PHOX2B exhibited a significant difference (Figure S2). In BM, there was a significant difference in the ΔCt values of the five genes between positive and negative specimens (BM morphology or flow cytometric MRD) according to the t-test (Figures S3,S4). ANOVA was performed to evaluate whether tumor burden made an independent contribution to NB5 ΔCt values. The relationship between NB5 ΔCt and flow cytometric MRD and BM morphology was analyzed. The results indicated that flow cytometric MRD was independently associated with a stronger NB5 genes expression level in both BM and PB (Table 4). Moreover, in the interaction terms between BM morphology and flow cytometric MRD, no significant differences in NB5 genes expression were observed.

Table 4
| Clinical disease comparison | BM NB5 assay ΔCt | PB NB5 assay ΔCt | |||
|---|---|---|---|---|---|
| ΔCt difference (SE) | P | ΔCt difference (SE) | P | ||
| Bone marrow morphology (negative vs. positive patients) | |||||
| CHGA | 14.365 (1.110) | 0.437 | 7.767 (2.188) | 0.856 | |
| DCX | 14.899 (0.071) | 0.164 | 7.142 (1.727) | 0.998 | |
| DDC | 13.691 (1.242) | 0.279 | 6.229 (1.889) | 0.546 | |
| PHOX2B | 15.518 (0.334) | 0.222 | 8.327 (2.168) | 0.773 | |
| TH | 13.215 (1.564) | 0.303 | 6.061 (1.617) | 0.817 | |
| MRD (flow cytometry) (negative vs. positive patients) | |||||
| CHGA | 14.743 (1.321) | <0.001 | 8.754 (2.546) | 0.040 | |
| DCX | 14.361 (1.392) | <0.001 | 7.665 (1.708) | 0.047 | |
| DDC | 13.605 (1.898) | <0.001 | 8.659 (1.982) | 0.006 | |
| PHOX2B | 14.997 (1.773) | <0.001 | 9.990 (3.073) | 0.014 | |
| TH | 13.381 (1.603) | <0.001 | 6.648 (1.574) | 0.014 | |
†, for main effects via ANOVA, ΔCt difference represents the difference between negative patients and positive patients of NB5 ΔCt. P<0.05 is a statistically significant difference. ANOVA, analysis of variance; NB5, five neuroblastoma genes (CHGA, DCX, DDC, PHOX2B, and TH); CHGA, chromogranin A; DCX, doublecortin; DDC, dopa decarboxylase; PHOX2B, paired-like homeobox 2b; TH, tyrosine hydroxylase; BM, bone marrow; PB, peripheral blood; SE, standard error; ΔCt, Δ threshold cycle; MRD, measurable residual disease.
Detection and analysis of NB5 ddPCR and qRT-PCR in BM, PB, and CSF
For different specimens (BM, PB, and CSF), we performed ddPCR on 24 specimens (20 patients). Based on previous testing of positive specimens, a threshold line of 0.010% (relative copy number) was established for ddPCR. The results, as shown in Table 5, revealed that the positive rates of ddPCR were consistent with qRT-PCR in BM (13/18, 72.222%) and CSF (1/1, 100.000%). However, in PB (2/5, 40.000%), the positive rate of ddPCR was higher than that of qRT-PCR (Table 5).
Table 5
| Method | Sample | Category description | N (%) |
|---|---|---|---|
| ddPCR | Bone marrow | Positive/total† | 13/18 (72.222) |
| Peripheral blood | Positive/total† | 2/5 (40.000) | |
| Cerebrospinal fluid | Positive/total† | 1/1 (100.000) | |
| qRT-PCR | Bone marrow | Positive/total† | 13/18 (72.222) |
| Peripheral blood | Positive/total† | 1/5 (20.000) | |
| Cerebrospinal fluid | Positive/total† | 1/1 (100.000) |
†, number of assessments. ddPCR, droplet digital polymerase chain reaction; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
The contribution of the NB5 genes to disease
A total of 38 specimens (BM, PB, and CSF) were detected positive as with qRT-PCR. Among the positive results, we assessed the frequency of the five genes in the positive specimens and used the χ2 test to determine if there were significant differences. The analysis revealed a significant difference (P=0.005) between the CHGA and TH genes in pairwise comparisons. No significant differences were observed in the pairwise comparisons of the other genes. Thus, based on qRT-PCR results, the contribution of the five genes was ranked as follows: CHGA ≈ DCX ≈ DDC ≈ PHOX2B and CHGA > TH (Table 6). In addition, we conducted the same analysis using ddPCR data but did not obtain results similar to those from qRT-PCR. This discrepancy may be related to the limited number of specimens used in the analysis (Table S4).
Table 6
| Gene | Positive† | Negative† | Total |
|---|---|---|---|
| CHGA | 33a (86.84) | 5a (13.16) | 38 (100.00) |
| DCX | 27a,b (71.05) | 11a,b (28.95) | 38 (100.00) |
| DDC | 31a,b (81.58) | 7a,b (18.42) | 38 (100.00) |
| PHOX2B | 30a,b (78.95) | 8a,b (21.05) | 38 (100.00) |
| TH | 21b (55.26) | 17b (44.74) | 38 (100.00) |
| Total | 142 (74.74) | 48 (25.26) | 190 (100.00) |
Data are shown as n (%). †, the letters “a” and “b” indicate whether there is a statistical difference; containing the same letters means there is no significant difference, otherwise there is a significant difference. qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; NB5, five neuroblastoma genes (CHGA, DCX, DDC, PHOX2B, and TH); CHGA, chromogranin A; DCX, doublecortin; DDC, dopa decarboxylase; PHOX2B, paired-like homeobox 2b; TH, tyrosine hydroxylase.
NB5 assay ΔCt and PFS
The analysis included time-dependent covariate analysis on ΔCt and PFS, with a Cox proportional hazards regression model was used to assess the hazard ratios (HRs) of different influencing factors. Our results showed that although there were no significant differences in flow cytometric MRD, BM morphology, or NB5 ΔCt for PFS, the grouping based on NB5 ΔCt demonstrated a tendency toward better prognostic stratification (Figure 4 and Table S5). In order to further evaluate the significance of NB5 ΔCt in prognostic stratification, additional analyses were conducted. We analyzed the PFS of patients without flow cytometric MRD detectable in correlation with NB5 ΔCt at two groups (ΔCt ≤18 and ΔCt >18). The average survival time in the positive group (NB5 ΔCt ≤18) of patients at initial diagnosis was 27.408±10.791 months (median, 21.000±3.914 months) compared with 35.961±3.084 months (median, 38.000±6.244 months) (P=0.034) in the negative group (NB5 ΔCt >18). Furthermore, the Cox analysis revealed that in flow cytometric MRD negative patients, the HR of the NB5 ΔCt positive group (ΔCt ≤18) was 3.046 times higher than that of the NB5 ΔCt negative group (ΔCt >18) (Figure 4D and Table S5). This result suggests that NB5 provides a more accurate assessment of disease progression and prognosis than does flow cytometric MRD and can be used as an independent factor to assess the role of clinical disease and prognosis.

Receiver operating characteristic (ROC) analysis was carried out for the ΔCt of the five genes. Additionally, we analyzed the geometric mean (GM) (GM Ct value of the five genes minus the GM of the housekeeping gene Ct, NB5 signature ΔCt) of the five genes and the other signatures (NB5 minus CHGA, NB5 minus DDC, NB5 minus DCX, NB5 minus PHOX2B, TH/DCX, PHOX2B/TH/DDC, PHOX2B/TH/DCX, and PHOX2B/TH), and a similar analysis was performed. Based on these results, the most influential single gene was found to be DCX, with an area under the curve (AUC) of 0.733. Its removal resulted in a 6.589% drop in the AUC (NB5 minus DCX). The results showed that the prediction of PFS by the DCX gene had the highest AUC (DCX > DDC > PHOX2B > CHGA > TH), and the AUC prediction result of the NB5 signature ΔCt was between DCX and DDC (Figure S5 and Table S6).
Discussion
Disease assessment is critical for therapeutic decisions and prognostic assessment. Several potential factors are associated with the risk stratification of high-risk NB, including clinical factors [e.g., age, serum lactate dehydrogenase (LDH), serum ferritin], host factors, tumor biology (e.g., MYCN amplification, genomic instability), disease response (e.g., MIBG scoring), and MRD monitoring (29). Among these numerous factors, rapid and accurate monitoring of MRD plays a crucial role in evaluating NB metastasis and residual disease, guiding treatment, and assessing prognosis. Previous studies have shown that the expression levels of CHGA, DCX, DDC, PHOX2B, and TH can be used alone or in combination as markers to guide micrometastasis (14,30).
CHGA is a member of the chromogranin/secretogranin family and is involved in encoding neurosecretory granules that promoting NB cell differentiation (31). The DCX gene encodes a protein belonging to the doublecortin family, which is specifically expressed in migrating neurons of the central and peripheral nervous systems. The expressed protein guides neuron migration by regulating microtubule organization and stability (32). DDC gene encodes a protein that catalyzes the formation of dopamine and is considered a sensitive marker for NB cells (33). The PHOX2B gene encodes a protein belonging to the paired family of homeodomain transcription factor, which is involved in neural maturation and differentiation. It is highly expressed in NB and has been identified as a specific marker for MRD in NB (34). The TH gene encodes a protein that functions as the rate-limiting enzyme in the synthesis of catecholamines and plays a key role in the physiology of adrenergic neurons. Given that catecholamines are predominantly produced by NB cells, TH plays a significant role in micrometastasis detection (35). Therefore, assessing the expression of these five genes is of great clinical value in NB detection.
qRT-PCR was utilized to detect three different specimens (BM, PB, CSF). The results indicated that all three specimens were capable of yielding positive results. In the CSF specimens, one patient in the consolidation therapy stage experienced seizure symptoms after radiotherapy, raising strong suspicion of central nervous system relapse. In the qRT-PCR analysis, four genes (DCX, DDC, PHOX2B, and CHGA) were found to be positive. However, after a few days of treatment, only three genes (DCX, PHOX2B, and CHGA) remained positive. Although the number of specimens was limited, the results strongly indicate the potential of applying qRT-PCR detection methods in different specimens (Table 5). In both ddPCR and qRT-PCR analyses, the positive results obtained from ddPCR were found to be 100% concordant with the results from qRT-PCR (Table 5). Interestingly, in PB specimens, ddPCR exhibited a higher positivity rate compared to qRT-PCR (with an additional positive detection). Concurrent assessment of BM MRD (flow cytometry) indicated a positive result (16.500%) for the same period in the patients. Based on this data, we speculate that for patients who are unable to provide a BM specimen, using ddPCR for PB diagnostic evaluation could be a viable strategy worth exploring. Nino et al. used ddPCR to detect MRD in PB stem cell grafts of patients with high-risk NB and found that MRD had an impact on the prognosis (36). Through the analysis of seven genes, they observed that patients with high gene expression had significantly lower event-free survival (EFS) (36). Our results, consistent with Nino’s findings, suggest that ddPCR holds greater potential for the application of MRD monitoring in NB.
Using individual genes for evaluating the results can enhance the sensitivity of the detection. However, to avoid false-positive results, we adopted a strategy inspired by previous studies (17,37,38) and compared the performance of the five individual genes with that of the NB5-signature approach (Figure S5 and Table S6). We also compared the results with other previously reported signatures (18,30,39,40). The results demonstrated that among all the different gene signature approaches, DCX alone consistently had the highest AUC (Figure S5 and Table S6). Furthermore, we observed that some patients showed positivity in only one gene (one case for TH, two cases for PHOX2B, and two cases for CHGA). Among these patients, the individual with TH positivity (flow cytometric MRD was negative) showed evidence of increased bone metabolism in the left scapula on imaging, suggestive of tumor bone metastasis. Remarkably, these patients demonstrated significant improvement in their condition after treatment. These findings further demonstrate that even when individual genes are positive, they can accurately assess the degree of disease progression.
In our study, a high correlation was observed between ΔCt values of the five genes in paired BM and PB specimens. Furthermore, NB5 ΔCt was detectable when BM morphology or MRD (flow cytometry) standard evaluations were negative, and these patients were found to have different degrees of disease progression or poor treatment effect in the later stage, which was consistent with the NB5 assay (Figure 2, Table 4).
The most notable results of this study are the correlation between NB5 expression levels in BM and PFS in patients with refractory or relapsed NB. This was especially evident in patients with negative for MRD according to flow cytometry. The data with undetected NB5 expression in BM was consistently associated with negative MRD results on flow cytometry and indicated a higher proportion of PFS. In contrast, among patients with MRD negative status on flow cytometry, those with NB5 positivity demonstrated poorer PFS according to the survival analysis and also had a higher HR compared to NB5-negative patients (Figure 4 and Table S5). For example, 1 patient was initially detected as positive for NB5 (DCX, DDC, PHOX2B, CHGA) but was negative for MRD on flow cytometry. After 6 months, a follow-up BM examination showed that NB5 (TH, DCX, DDC, PHOX2B, CHGA) remained positive. At this time, both flow cytometric MRD and BM morphology showed positive results. Another patient was negative on both the first and second NB5 assay, and was in the maintenance treatment phase in a CR state. In the third NB5 assay, all five genes were found to be positive, but BM morphology and flow cytometric MRD monitoring were all negative. Additionally, imaging examinations did not reveal any signs of recurrence. Close follow-up monitoring was conducted, and a re-examination half a month later showed MIBG uptake near the left renal hilum, the bilateral mandibular branches, bilateral the iliac bones, and a focally in a lesion on the right ischium, suggestive of bone metastasis. The Curie score was 5 points, indicating widespread relapse. These two cases clearly illustrate that BM NB5 detection is more sensitive and more accurate than is BM flow cytometric MRD in reflecting the BM conditions of patients with NB. In addition, when the NB5 detection result change from negative to positive, this could predict NB recurrence earlier, and early intervention and treatment can greatly improve the final prognosis of patients.
We further found that BM NB5 detection has advantages over BM flow cytometric MRD detection in assessing treatment effect. In a 3-year-old girl, the first BM NB5 detection was positive for TH and DDC, and flow cytometric MRD (0.010%) was also positive. After induction therapy was continued, the second BM examination showed that flow cytometric MRD was negative. However, TH and DDC genes were still detected, and the patient still had multiple lesions in the bone. At the time of writing, the patient is in a PR state. Our data suggest that for patients with relapsed/refractory NB, even in cases where MRD is clearly negative, NB5 results can still accurately predict PFS. The expression of NB5 in BM enables early risk stratification for patients with relapsed or refractory NB before and during treatment. Several studies have explored the use of different genes to the assess survival rate in NB. Viprey et al. investigated the expression levels of PHOX2B, TH, and DCX in BM and found that these expression levels could be used to predict EFS and OS, as well as monitor treatment response (30). Additionally, Marachelian et al. used CHGA, DCX, DDC, PHOX2B, and TH to assess expression levels and their significant correlation with PFS (14). Despite the small number of false positives and false negatives, multiple larger multicenter studies, the inclusion of additional clinical features, and use of diverse detection methods are required to comprehensively evaluate and explore the application value of these five genes in NB.
Conclusions
Our findings demonstrate that NB5 detection in the BM of patients with NB is more sensitive and provides a more accurate reflection of the BM status of patients with NB as compared to BM flow cytometric MRD. It can precisely assess prognosis and prevent tumor recurrence in patients with NB. Furthermore, BM NB5 detection exhibited higher sensitivity and accuracy than did PB NB5 detection. BM NB5 detection may also serve as an independent prognostic factor for patients with relapsed or refractory NB.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-545/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-545/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-545/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-545/coif). H.L. is from Shanghai Cinopath Medical Laboratory Co., Ltd., Shanghai, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Medical Ethics Committee of Hunan Provincial People’s Hospital (approval No. 2023-106). Informed consent was obtained from all the patients’ legal guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mlakar V, Morel E, Mlakar SJ, et al. A review of the biological and clinical implications of RAS-MAPK pathway alterations in neuroblastoma. J Exp Clin Cancer Res 2021;40:189. [Crossref] [PubMed]
- Teitz T, Inoue M, Valentine MB, et al. Th-MYCN mice with caspase-8 deficiency develop advanced neuroblastoma with bone marrow metastasis. Cancer Res 2013;73:4086-97. [Crossref] [PubMed]
- Ambros PF, Ambros IM, Brodeur GM, et al. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer 2009;100:1471-82. [Crossref] [PubMed]
- London WB, Castel V, Monclair T, et al. Clinical and biologic features predictive of survival after relapse of neuroblastoma: a report from the International Neuroblastoma Risk Group project. J Clin Oncol 2011;29:3286-92. [Crossref] [PubMed]
- Irwin MS, Naranjo A, Zhang FF, et al. Revised Neuroblastoma Risk Classification System: A Report From the Children's Oncology Group. J Clin Oncol 2021;39:3229-41. [Crossref] [PubMed]
- Bergfeld SA, Blavier L, DeClerck YA. Bone marrow-derived mesenchymal stromal cells promote survival and drug resistance in tumor cells. Mol Cancer Ther 2014;13:962-75. [Crossref] [PubMed]
- Fang H, Declerck YA. Targeting the tumor microenvironment: from understanding pathways to effective clinical trials. Cancer Res 2013;73:4965-77. [Crossref] [PubMed]
- Yu AL, Gilman AL, Ozkaynak MF, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med 2010;363:1324-34. [Crossref] [PubMed]
- Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279-84. [Crossref] [PubMed]
- Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015;526:700-4. [Crossref] [PubMed]
- de Mestier L, Dromain C, d'Assignies G, et al. Evaluating digestive neuroendocrine tumor progression and therapeutic responses in the era of targeted therapies: state of the art. Endocr Relat Cancer 2014;21:R105-20. [Crossref] [PubMed]
- Cheung NK, Heller G, Kushner BH, et al. Detection of metastatic neuroblastoma in bone marrow: when is routine marrow histology insensitive? J Clin Oncol 1997;15:2807-17. [Crossref] [PubMed]
- Manenq C, Lesesve JF, Dreumont N, et al. Combined use of multiparametric flow cytometry and cytomorphology to enhance detection of neuroblastoma metastatic cells in bone marrow. Int J Lab Hematol 2020;42:52-60. [Crossref] [PubMed]
- Marachelian A, Villablanca JG, Liu CW, et al. Expression of Five Neuroblastoma Genes in Bone Marrow or Blood of Patients with Relapsed/Refractory Neuroblastoma Provides a New Biomarker for Disease and Prognosis. Clin Cancer Res 2017;23:5374-83. [Crossref] [PubMed]
- Grèze V, Brugnon F, Chambon F, et al. Highly sensitive assessment of neuroblastoma minimal residual disease in ovarian tissue using RT-qPCR-A strategy for improving the safety of fertility restoration. Pediatr Blood Cancer 2017; [Crossref]
- Stutterheim J, Zappeij-Kannegieter L, Versteeg R, et al. The prognostic value of fast molecular response of marrow disease in patients aged over 1 year with stage 4 neuroblastoma. Eur J Cancer 2011;47:1193-202. [Crossref] [PubMed]
- Thwin KKM, Ishida T, Uemura S, et al. Level of Seven Neuroblastoma-Associated mRNAs Detected by Droplet Digital PCR Is Associated with Tumor Relapse/Regrowth of High-Risk Neuroblastoma Patients. J Mol Diagn 2020;22:236-46. [Crossref] [PubMed]
- Yáñez Y, Hervás D, Grau E, et al. TH and DCX mRNAs in peripheral blood and bone marrow predict outcome in metastatic neuroblastoma patients. J Cancer Res Clin Oncol 2016;142:573-80. [Crossref] [PubMed]
- Cheung IY, Cheung NK. Quantitation of marrow disease in neuroblastoma by real-time reverse transcription-PCR. Clin Cancer Res 2001;7:1698-705.
- Cheung IY, Feng Y, Vickers A, et al. Cyclin D1, a novel molecular marker of minimal residual disease, in metastatic neuroblastoma. J Mol Diagn 2007;9:237-41. [Crossref] [PubMed]
- Stutterheim J, Gerritsen A, Zappeij-Kannegieter L, et al. Detecting minimal residual disease in neuroblastoma: the superiority of a panel of real-time quantitative PCR markers. Clin Chem 2009;55:1316-26. [Crossref] [PubMed]
- Viprey VF, Lastowska MA, Corrias MV, et al. Minimal disease monitoring by QRT-PCR: guidelines for identification and systematic validation of molecular markers prior to evaluation in prospective clinical trials. J Pathol 2008;216:245-52. [Crossref] [PubMed]
- Kreissman SG, Seeger RC, Matthay KK, et al. Purged versus non-purged peripheral blood stem-cell transplantation for high-risk neuroblastoma (COG A3973): a randomised phase 3 trial. Lancet Oncol 2013;14:999-1008. [Crossref] [PubMed]
- Tang J, Lu H, Yang Z, et al. Associations between WTAP gene polymorphisms and neuroblastoma susceptibility in Chinese children. Transl Pediatr 2021;10:146-52. [Crossref] [PubMed]
- Pérez-García MJ, Segura MF. Maintainingexcellent outcomes: the impact of age cutoff reclassificationon reduced therapy for neuroblastoma patients. Transl Pediatr 2023;12:1926-30.
- Druy AE, Shorikov EV, Tsaur GA, et al. Prospective investigation of applicability and the prognostic significance of bone marrow involvement in patients with neuroblastoma detected by quantitative reverse transcription PCR. Pediatr Blood Cancer 2018;65:e27354. [Crossref] [PubMed]
- Huggett JF, Foy CA, Benes V, et al. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem 2013;59:892-902. [Crossref] [PubMed]
- Dixon WJ, Massey FJ. Introduction to statistical analysis. 3rd ed. New York, NY: McGraw-Hill; 1968.
- Morgenstern DA, Bagatell R, Cohn SL, et al. The challenge of defining "ultra-high-risk" neuroblastoma. Pediatr Blood Cancer 2019;66:e27556. [Crossref] [PubMed]
- Viprey VF, Gregory WM, Corrias MV, et al. Neuroblastoma mRNAs predict outcome in children with stage 4 neuroblastoma: a European HR-NBL1/SIOPEN study. J Clin Oncol 2014;32:1074-83. [Crossref] [PubMed]
- Gaetano C, Manni I, Bossi G, et al. Retinoic acid and cAMP differentially regulate human chromogranin A promoter activity during differentiation of neuroblastoma cells. Eur J Cancer 1995;31A:447-52. [Crossref] [PubMed]
- Oltra S, Martinez F, Orellana C, et al. The doublecortin gene, a new molecular marker to detect minimal residual disease in neuroblastoma. Diagn Mol Pathol 2005;14:53-7. [Crossref] [PubMed]
- Bozzi F, Luksch R, Collini P, et al. Molecular detection of dopamine decarboxylase expression by means of reverse transcriptase and polymerase chain reaction in bone marrow and peripheral blood: utility as a tumor marker for neuroblastoma. Diagn Mol Pathol 2004;13:135-43. [Crossref] [PubMed]
- Bachetti T, Di Zanni E, Ravazzolo R, et al. miR-204 mediates post-transcriptional down-regulation of PHOX2B gene expression in neuroblastoma cells. Biochim Biophys Acta 2015;1849:1057-65. [Crossref] [PubMed]
- Burchill SA, Lewis IJ, Abrams KR, et al. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mRNA are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J Clin Oncol 2001;19:1795-801. [Crossref] [PubMed]
- Nino N, Ishida T, Nakatani N, et al. Minimal residual disease detected by droplet digital PCR in peripheral blood stem cell grafts has a prognostic impact on high-risk neuroblastoma patients. Heliyon 2022;8:e10978. [Crossref] [PubMed]
- Batchu S. Immunological landscape of Neuroblastoma and its clinical significance. Cancer Treat Res Commun 2021;26:100274. [Crossref] [PubMed]
- Asgharzadeh S, Marachelian A, Villablanca JG, et al. Expression of neuroblastoma-related genes in bone marrow at end of high-risk neuroblastoma therapy. Pediatr Blood Cancer 2022;69:e29719. [Crossref] [PubMed]
- Cheung NK, Ostrovnaya I, Kuk D, et al. Bone marrow minimal residual disease was an early response marker and a consistent independent predictor of survival after anti-GD2 immunotherapy. J Clin Oncol 2015;33:755-63. [Crossref] [PubMed]
- van Wezel EM, Decarolis B, Stutterheim J, et al. Neuroblastoma messenger RNA is frequently detected in bone marrow at diagnosis of localised neuroblastoma patients. Eur J Cancer 2016;54:149-58. [Crossref] [PubMed]


