
Application of neural networks in prenatal diagnosis of atrioventricular septal defect
Highlight box
Key findings
• U-net, Link-net, and MA-net could detect landmarks on fetal echocardiography (FE) and make the measurements accurately.
What is known and what is new?
• The atrial length to ventricular length ratio, acquired on the four-chamber view in end-diastole, could be used to identify atrioventricular septal defect prenatally. Automatic detection and assessment of landmarks, an important and active topic in the field of medical image processing research, has not been applied in the FE area so far.
• U-net, Link-net, and MA-net could detect landmarks on FE and make the measurements accurately.
What is the implication, and what should change now?
• Landmarks detection technology could be used to make quantitative measurements for cardiac structures on FE. That means landmarks detection technology could be used to make prenatal screening or diagnosis for cardiovascular malformations in which the ventricles or large arteries are proportional abnormally.
Introduction
Atrioventricular septal defect (AVSD) is featured with the complete or partial absence of the atrioventricular septum and therefore presenting a common atrioventricular junction (1,2). On fetal echocardiography (FE), careful examination of the crisscross, atrioventricular annulus, and atrioventricular valves is conducive to the diagnosis (3). But this depends heavily on the sonographers’ experience as well as the geographical and socioeconomic factors (3). Recently, Aldridge (4) investigated the prenatal detection rate for 14 kinds of congenital anomalies in the UK and found about 40% of AVSD fetuses could not be screened out even if the National Health Service Fetal Anomaly Screening Program was introduced 13 years ago. In 2004, Machlitt (5) proposed a simple parameter, the atrial length (AL) to ventricular length (VL) ratio (AVLR), acquired on the four-chamber view in end-diastole, to identify AVSD prenatally and the detection rate could reach 86%. And in 2023, Zhou et al. (6) further validated the diagnostic ability of AVLR in the prenatal examination for AVSD through a relatively large-scale study. However, due to busy work for fetal heart detection, few examiners would like to remember one more value.
Neural networks, one way to implement artificial intelligence and designed to focus on the algorithms to learn, perform and complete various missions and processes in a human way, have been widely developed in the medical field so far (7,8). “Landmarks” are defined as some points or curves with specific characteristics and corresponding relationships in location and topology in the sense of medical anatomy (9). Automatic detection and assessment of landmarks, one of the Convolutional Neural Network algorithms, is an important and active topic in the field of medical image processing research (10). In the echocardiographic field, landmark detection was used to estimate the ventricular function (11), detect the prolapse location of the mitral valve (12), and detect the images’ border automatically (13). However, there is no relevant literature about landmarks detection in the field of FE. Congenital cardiovascular defects affect the outcomes of newborns or infants and the quality of life of individuals and families (14). In addition, Vena and the colleagues (15) show that obstructive lesions of the left-side and right-side cardiac system are associated with fetal neurodevelopmental abnormalities. Therefore, it is important to make prenatal detection of congenital heart disease, which could stratify the risk of the disease, provide a basis for management and decision-making prenatally as well as reasonable treatment and intervention postnatally, improve the treatment effects, reduce the mortality, and improve the quality of life of children.
Therefore, the purpose of this study was to explore whether the landmarks on FE could be detected correctly by three state-of-the-art neural networks including U-net (16), MA-net (17), and Link-net (18) and whether AVLR could be used to make the prenatal diagnosis of AVSD. We present this article in accordance with the TRIPOD reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-394/rc).
Methods
Data collection
This was an observational study. Two hundred and seventy-five fetuses were consecutively included from the Maternal-Fetal Consultation Center of Congenital Heart Disease, Beijing Anzhen Hospital from December 2017 to January 2022. Among these fetuses, 117 were normal, 53 were diagnosed with AVSD, 29 were hypoplastic right heart syndrome (HRHS), 45 were ventricular septal defects (VSD) cases, and 31 were hypoplastic left heart syndrome (HLHS). AVSD fetuses were grouped as the study group and the normal fetuses were the control group. The HRHS, VSD, and HLHS fetuses, were grouped as the differential diagnosis group. At the time of the ultrasound examination, the mean age of pregnant women was 28.6±4.5 years old, and the mean gestational age for the fetuses were 24.4±4.1 weeks. Table 1 shows the details of general information in 275 fetuses.
Table 1
| Characteristics | Normal | AVSD | HRHS | VSD | HLHS |
|---|---|---|---|---|---|
| Numbers | 117 | 53 | 29 | 45 | 31 |
| Gestational weeks | 25.4±4.1 | 24.3±3.3 | 23.5±2.6 | 24.1±3.2 | 25.2±3.6 |
| Age of the pregnant (years) | 29.5±5.1 | 28.4±3.5 | 28.3±3.6 | 29.2±4.6 | 28.5±3.1 |
All data of the gestational weeks and the age of the pregnant were presented as means ± standard deviations. AVSD, atrioventricular septal defect; HRHS, hypoplastic right heart syndrome; HLHS, hypoplastic left heart syndrome; VSD, ventricular septal defect.
Voluson E10-C1-5-D (GE Healthcare, Zipf, Austria) and Voluson E8-RAB 4-8 (GE Healthcare, Milwaukee, Wisconsin, USA) equipped with the 2 to 4 MHz transducer, and the Aloka 10 UST-9130 (Aloka, Tokyo, Japan) equipped with the 3 to 6 MHz transducer were used for all FE examinations. Standard images including the four-chamber, outflow tract, three-vessel or three-vessel trachea, and the long axis view of aortic arch as well as the color and pulse doppler were analyzed according to the segmental analysis method (19). All images were stored in the database finally. In our research, the final diagnosis for all fetuses was confirmed by the postnatal results on echocardiography, the autopsy, or confirmed by two senior doctors with at least 10 years’ experience in FE. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ethics committee of Beijing Anzhen Hospital approved this study (No. 2020016X) and the informed consent for autopsy was obtained from the parents.
Dataset preparing
Due to the fact that three AVSD fetuses underwent the second FE, 56 datasets were included in the AVSD group ultimately. A total of 278 datasets in 4DV format were imported into the ultrasound machines to convert to JPG, MOV, or AVI formats which were prepared for the neural network models. All readable format images were imported into the labeled system to prepare for the landmarks labeling. As AVLR was acquired on the four-chamber view in end-diastole, the four-chamber videos were finally selected and used.
The details were as follows: firstly, select one frame in the end diastole, and in this frame, the ventricles were the largest with the closed atrioventricular valves. Secondly, a junior physician with 3–5 years of experience in FE labeled seven landmarks. In normal fetuses and differential diagnosis groups, sequence of seven landmarks is shown in Figure 1A-1D. In the AVSD group, the only difference was the landmark 2 which was located at the closed common atrioventricular valves instead (Figure 1E,1F). Among these landmarks, landmarks of 1, 2, 3 were used to measure AL, VL and calculate AVLR, and the 4 to 7 were used to verify the detective capacity of landmarks for neural networks. The average time for labeling seven landmarks in an image took approximately 7 seconds. Thirdly, 278 labeled images were reviewed and the inaccurate landmarks were corrected by a chief physician with over 10 years of experience in FE. Eventually, 278 labeled frames were used as the datasets. Table 2 shows the details of the training and test sets in the normal, AVSD, HRHS, VSD, and HLHS groups.

Table 2
| Groups | Training set | Test set | Total |
|---|---|---|---|
| Normal | 96 | 21 | 117 |
| AVSD | 42 | 14 | 56 |
| HRHS | 23 | 6 | 29 |
| VSD | 36 | 9 | 45 |
| HLHS | 25 | 6 | 31 |
| Total | 222 | 56 | 278 |
AVSD, atrioventricular septal defect; HRHS, hypoplastic right heart syndrome; HLHS, hypoplastic left heart syndrome; VSD, ventricular septal defect.
Data processing
Data processing, realized by using Python, mainly involved establishing the standard datasets to eliminate errors originating from different ultrasound machines and operators as much as possible. During this process, all labeled frames were exported in JPG format for further analysis. The details were: firstly, due to there being 876×1,280, 720×960, 880×1,264, and other pixels for the original images, it was necessary to standardize all images into 384×512 pixels to simplify the training and testing process for models. And then the patients’ privacy area was removed and the final pixels for each image were 340×450. Finally, to unify the grayscale of all images to the same interval, the images were normalized according to the formula: , where max is the maximum value of the sample data, and min is the minimum value of the sample data.
Model building
The heatmap, generated by the landmarks labeled by doctors, was used as the center, and the probability value of the position for each landmark was defined as 1. Taking the landmarks as the center, the Gaussian function was used to gradually decrease the probability value outward. The semantic segmentation models were applied as the landmark prediction networks. The original images were used as the input images for the models and seven channels were used as the outputs, with each output channel predicting a specific landmark. The detection performance of three networks including the U-net (16), MA-net (17), and Link-net (18) was compared in this study.
Training procedure
During the training process, the number of batch images was set to 8. Taking mean squared error (MSE) as the loss function, the learning rate was set to 0.0000009. The RMSprop optimizer was used and the gradient attenuation weight was set to 0.0001. By observing the output loss value, it was found that the networks were basically stable after 30 rounds of training, then the network with a smaller value was selected as the test model. In the training process, the networks were adjusted to the training mode, to optimize the intermediate parameters such as the average of batch standardization. In the process of reasoning, the architectures were adjusted to the eval mod; freeze the parameters of the networks so that the parameters would not be modified in the process of reasoning. In our study, the processes of training and reasoning were operated on the NVIDIA Tesla V100 graphics card.
Statistical analysis
All statistical analyses were conducted using IBM SPSS 23.0 version and Med-calc 19.0 version statistical software. Bland-Altman consistency analysis was used to verify the consistency of measurement values between three models and expert labels. Continuous variables were shown as a mean ± standard deviation (SD). Unpaired Student’s t-test was applied for comparison between two groups, and the one-way analysis of variance (ANOVA) test was used among three or more groups. A two-sided test was utilized in all tests, and the P value <0.05 was considered the statistical difference. To differentiate normal and AVSD, the cut-off value of the AVLR was acquired. The sensitivity, specificity, Youden index, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (PLR), negative likelihood ratio (NLR), and the area under the receiver operating characteristic (ROC) curves (AUC), as well as the 95% confidence intervals (CIs), were also calculated.
Results
There were 117 normal, 29 HRHS, 45 VSD, and 31 HLHS and 53 AVSD fetuses included in this study. In 53 AVSD fetuses, 31 were complete, 12 were partial, and 10 were transitional. And there were 30 isolated fetuses, 13 cases combined with left ventricular outflow tract obstruction, and 10 cases combined with the right ventricular outflow tract obstruction in this group. In the testing group, the numbers of the complete, partial, and transitional subgroups were 10, 2, and 2 separately. The statistical results showed the difference of AVLR among the complete, partial, and transitional subgroups was not significant (P>0.05), but was significant between the normal and the three subgroups (P<0.05) (Figure 2, Table 3).
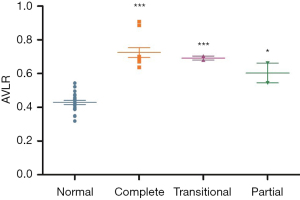
Table 3
| Subgroups | AVLR |
|---|---|
| Normal | 0.42±0.06 |
| Complete | 0.72±0.13*** |
| Transitional | 0.67±0.01*** |
| Partial | 0.60±0.08* |
All values were shown as means ± standard deviations. *, there was statistical difference between the partial AVSD and the normal fetuses with the P value <0.05; ***, there were statistical differences between the AVSD subgroups and the normal fetuses with the P value <0.0001. AVLR, atrial length to ventricular length ratio; AVSD, atrioventricular septal defect.
Comparisons of performance among three state-of-the-art networks and the manual
Figure 3 shows the difference between the landmarks labeled by the experts and detected by three models. All values were expressed by the pixel distance and were demonstrated by values on two axes, X and Y (X, Y). Table 4 demonstrates all coordinates labeled by the experts and detected by three models as well as their localization errors. The results showed U-net, Link-net, and MA-net could precisely detect all 7 landmarks, and the localization error on X and Y axis for seven landmarks were in the range of 0.09 and 0.13 respectively for the three networks.

Table 4
| Landmarks and the localization error | Manual (X, Y) | U-net (X, Y) | Link-net (X, Y) | MA-net (X, Y) |
|---|---|---|---|---|
| Landmark 1 | 236.848, 141.030 | 236.935, 141.028 | 236.936, 141.028 | 236.936, 141.028 |
| Localization error | – | 0.087, 0.002 | 0.087, 0.002 | 0.087, 0.002 |
| Landmark 2 | 226.363, 159.303 | 226.406, 159.178 | 226.406, 159.178 | 226.406, 159.179 |
| Localization error | – | 0.042, 0.124 | 0.042, 0.124 | 0.042, 0.124 |
| Landmark 3 | 219.667, 168.242 | 219.586, 168.278 | 219.586, 168.278 | 219.586, 168.278 |
| Localization error | – | 0.080, 0.035 | 0.080, 0.035 | 0.080, 0.035 |
| Landmark 4 | 227.121, 177.363 | 227.058, 177.332 | 227.058, 177.332 | 227.058, 177.332 |
| Localization error | – | 0.063, 0.031 | 0.063, 0.031 | 0.063, 0.031 |
| Landmark 5 | 227.333, 175.061 | 227.273, 175.061 | 227.273, 175.061 | 227.273, 175.061 |
| Localization error | – | 0.060, 0.000 | 0.060, 0.000 | 0.060, 0.000 |
| Landmark 6 | 223.394, 142.121 | 223.398, 142.008 | 223.398, 142.008 | 223.398, 142.008 |
| Localization error | – | 0.004, 0.113 | 0.004, 0.113 | 0.004, 0.113 |
| Landmark 7 | 222.212, 134.333 | 222.205, 134.327 | 222.205, 134.327 | 222.205, 134.327 |
| Localization error | – | 0.008, 0.006 | 0.008, 0.006 | 0.008, 0.006 |
All data were expressed in means.
The results of this study showed three networks could measure the pixel distance of VL and AL accurately (Figure 4), and the measured error in AL and VL were 0.12 and 0.01 separately (Table 5).

Table 5
| Models | VL | AL | Measurement error | |
|---|---|---|---|---|
| VL | AL | |||
| Manual | 93.06±3.12 | 45.46±2.07 | – | – |
| U-net | 93.07±3.12 | 45.35±2.06 | 0.01±0.07 | 0.12±0.07 |
| Link-net | 93.07±3.12 | 45.35±2.06 | 0.01±0.07 | 0.12±0.07 |
| MA-net | 93.07±3.12 | 45.35±2.06 | 0.01±0.07 | 0.12±0.07 |
All values were presented as means ± standard deviations. AL, atrial length; VL, ventricular length.
AVLR in normal, AVSD, HRHS, VSD, and HLHS groups are shown in Table 6. The average time in the process from landmark detection, measurements, to the AVLR calculation were 0.0385, 0.04497, and 0.0393 second for U-net, MA-net, and Link-net respectively. This study showed AVLR calculated by the manual labeled and the three networks in AVSD fetuses were significantly larger than the other four groups (P<0.0001) (Figure 5), and there was no statistical difference among the normal, HRHS, VSD, and HLHS groups (P>0.05) (Figure 5).
Table 6
| Models | Normal group | AVSD group | HRHS group | VSD group | HLHS group |
|---|---|---|---|---|---|
| Manual | 0.42±0.06 | 0.70±0.09*** | 0.41±0.07 | 0.43±0.06 | 0.45±0.08 |
| U-net | 0.43±0.06 | 0.70±0.09*** | 0.41±0.06 | 0.43±0.06 | 0.45±0.07 |
| Link-net | 0.43±0.06 | 0.70±0.09*** | 0.40±0.06 | 0.43±0.05 | 0.45±0.08 |
| MA-net | 0.43±0.06 | 0.69±0.10*** | 0.39±0.06 | 0.43±0.06 | 0.44±0.08 |
***, there were statistical differences between the AVSD and the other four groups with P value <0.0001. AVLR, atrial length to ventricular length ratio; AVSD, atrioventricular septal defect; HRHS, hypoplastic right heart syndrome; HLHS, Hypoplastic left heart syndrome; VSD, ventricular septal defect.

The diagnostic effect for neural networks
This study showed the area under ROC curve (AUC) acquired from three neural networks could reach 0.992 (0.968–1.000), which was the same as the expert labeled (Figure 6). And the diagnostic accuracy, sensitivity, and specificity were 0.941, 100%, and 94.12% respectively. If 0.63 was selected as a cutoff value, the sensitivity for prenatal detection of AVSD fetuses was more than 90% within the false positive rate of 5%; and when 0.53 was chosen, all AVSD fetuses could be detected within the false positive rate of 6%. The AUC, sensitivity, specificity, Youden index, PLR, NLR, PPV, NPV, and their 95% CI are demonstrated in Table 7.
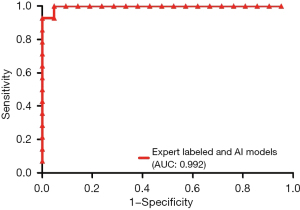
Table 7
| Parameters | Manual | U-net | Link-net | MA-net |
|---|---|---|---|---|
| AUC (95% CI) | 0.992 (0.968–1.000) | 0.992 (0.968–1.000) | 0.992 (0.968–1.000) | 0.992 (0.968–1.000) |
| P value | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Youden index | 0.941 | 0.941 | 0.941 | 0.941 |
| Sensitivity (95% CI) (%) | 100.00 (59.00–100.00) | 100.00 (59.00–100.00) | 100.00 (59.00–100.00) | 100.00 (59.00–100.00) |
| Specificity (95% CI) (%) | 94.12 (71.30–99.90) | 94.12 (71.30–99.90) | 94.12 (71.30–99.90) | 94.12 (71.30–99.90) |
| PLR (95% CI) (%) | 17.00 (2.54–113.83) | 17.00 (2.54–113.83) | 17.00 (2.54–113.83) | 17.00 (2.54–113.83) |
| NLR (95% CI) (%) | 0 (–) | 0 (–) | 0 (–) | 0 (–) |
| PPV (95% CI) (%) | 87.50 (51.1–97.9) | 87.50 (51.1–97.9) | 87.50 (51.1–97.9) | 87.50 (51.1–97.9) |
| NPV (95% CI) (%) | 100.00 (–) | 100.00 (–) | 100.00 (–) | 100.00 (–) |
AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value; PLR, positive likelihood ratio; NLR, negative likelihood ratio; CI, confidence interval; AVSD, atrioventricular septal defect.
Discussion
This study showed the performance of U-net (16), MA-net (17), and Link-net (18) could achieve the expert level in landmarks detection and cardiac measurement. AVLR acquired by landmarks detection could be used an effective parameter in the prenatal diagnosis of AVSD. As far as we know, this was the first time to apply landmark detection methods on FE.
AVSD occupies approximately 7.0% (20) of all congenital cardiac diseases and the complete subgroup is the most common (21). Our research revealed more than half (58%) AVSD fetuses were the complete, and this was consistent with Miller (21). Even though 80% (22) AVSD are with no other cardiac malformations, more than 45% (23) of AVSD combined with Down syndrome. In addition, other combinations including the outflow tract obstruction and the heterotaxia syndrome could make a great influence on the prognosis (24,25). Consequently, identifying AVSD timely is significant because it involves perinatal consultation, intrapartum and neonatal care, treatment options, survival time, as well as quality of life.
To date, artificial intelligence has been widely used in the FE area, and it is expected to improve the congenital cardiac abnormality detection rate by combining with traditional methods (10,11). In 2023, Veronese (26) used the fetal intelligent navigation echocardiography technology to realize the identification of AVSD, but it requires experienced sonographers to identify the views and the anatomical structures. Recently, the prenatal diagnostic effectiveness of AVLR was further proved by our team based on big data technology (6). Under normal conditions, due to the exitance of crisscross, the anterior leaflet of the mitral valve and septal leaflet of the tricuspid valve attach to the ventricular septum (27,28). AVSD is featured with the complete or partial absence of atrioventricular septum, resulting in the displacement of the atrioventricular valvular attachment and leading to a shortened inlet (29). Based on the anatomical characteristics of AVSD, this study realized the prenatal diagnosis and the differential diagnosis by detecting three landmarks. The results demonstrated the statistical difference of AVLR in the AVSD subgroups was not obvious but greatly higher than the normal and differential diagnostic fetuses. And this was consistent with the previous studies (5,6).
Three state-of-the-art neural networks, U-net (16), MA-net (17), and Link-net (18), were used for landmarks detection in our study. U-net (16) uses the encoder-decoder structure to construct outputs of the same size as the original images. It establishes a one-to-one correspondence between the outputs and the original images at the pixel level, and the outputs represent the probability of a certain category of pixels at that location. This network could directly splice the low-order semantic information with high-order features through means of jumping links. Therefore, it accepts the returned gradient information, thereby the model’s learning of semantic features is enhanced. And eventually, it reuses the features extracted during the up-sampling process and effectively improves the accuracy of up-sampling features. MA-net (17), a multi-scale attention net, is introduced the self-attention mechanism. This model could integrate the local information and the relevant global dependencies adaptively and then captures rich contextual dependencies. It could fuse the channel dependencies of high-level and low-level features by the sum method and obtain rich semantic information of multi-scale characteristics to improve the performance by using the attention mechanism. Link-net (18) connects each encoder and decoder and bypasses the input features of each encoder layer to the output of the corresponding decoder. By this way, it could recover the spatial information lost in the decoder and its up-sampling operations. In addition, due to the decoder sharing the knowledge learned by the encoder at each layer, the parameters used by the decoder are fewer. Our results showed the effectiveness of landmarks detection and the cardiac measurement in three models was accurate compared to the manual labeled.
The results in this study suggested the same diagnostic effectiveness of AVLR both in the expert labeled and three models. That may be related to the insufficient number of cases in the test set, which plays a crucial role in the process of evaluating diagnostic capability and selecting cutoff values. In addition, accurate detective effectiveness of three neural networks may be another important reason.
There were some limitations in this study. On the one hand, due to the small sample size of the test set, there might exist bias in selecting cutoff values. On the other hand, because images in readable format were used in this study, only the pixel distance of AL and VL instead of the specific measurement values was obtained. In future research, we will increase the sample size to minimize the bias as far as possible. In addition, The DICOM format images will be applied for specific cardiac measurements rather than pixel distance.
Conclusions
Landmark detection was an applicative method for prenatal diagnosis of AVSD. U-net, Link-net, and MA-net could detect landmarks and make the measurements accurately and rapidly on FE. The diagnostic effect of AVLR, calculated by three networks, was pretty good in prenatal diagnosis of AVSD.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-394/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-394/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-394/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-394/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ethics committee of Beijing Anzhen Hospital approved this study (No. 2020016X) and the informed consent for autopsy was obtained from the parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Słodki M, Soroka M, Rizzo G, et al. Prenatal Atrioventricular Septal Defect (AVSD) as a planned congenital heart disease with different outcome depending on the presence of the coexisting extracardiac abnormalities (ECA) and/or malformations (ECM). J Matern Fetal Neonatal Med 2020;33:2635-41. [Crossref] [PubMed]
- Calkoen EE, Hazekamp MG, Blom NA, et al. Atrioventricular septal defect: From embryonic development to long-term follow-up. Int J Cardiol 2016;202:784-95. [Crossref] [PubMed]
- Pan F, Li J, Lou H, et al. Geographical and Socioeconomic Factors Influence the Birth Prevalence of Congenital Heart Disease: A Population-based Cross-sectional Study in Eastern China. Curr Probl Cardiol 2022;47:101341. [Crossref] [PubMed]
- Aldridge N, Pandya P, Rankin J, et al. Detection rates of a national fetal anomaly screening programme: A national cohort study. BJOG 2023;130:51-8. [Crossref] [PubMed]
- Machlitt A, Heling KS, Chaoui R. Increased cardiac atrial-to-ventricular length ratio in the fetal four-chamber view: a new marker for atrioventricular septal defects. Ultrasound Obstet Gynecol 2004;24:618-22. [Crossref] [PubMed]
- Zhou X, Yang T, Zhang Y, et al. Quantitative Parameters Analysis for Prenatally Echocardiographic Diagnosis of Atrioventricular Septal Defects. Congenital Heart Disease 2023;18:387-97.
- Fiorentino MC, Villani FP, Di Cosmo M, et al. A review on deep-learning algorithms for fetal ultrasound-image analysis. Med Image Anal 2023;83:102629. [Crossref] [PubMed]
- Xiao S, Zhang J, Zhu Y, et al. Application and Progress of Artificial Intelligence in Fetal Ultrasound. J Clin Med 2023;12:3298. [Crossref] [PubMed]
- Hanaoka S, Shimizu A, Nemoto M, et al. Automatic detection of over 100 anatomical landmarks in medical CT images: A framework with independent detectors and combinatorial optimization. Med Image Anal 2017;35:192-214. [Crossref] [PubMed]
- Zhou GQ, Miao J, Yang X, et al. Learn Fine-Grained Adaptive Loss for Multiple Anatomical Landmark Detection in Medical Images. IEEE J Biomed Health Inform 2021;25:3854-64. [Crossref] [PubMed]
- Wang Z, Shi J, Hao X, et al. Simultaneous Right Ventricle End-diastolic and End-systolic Frame Identification and Landmark Detection on Echocardiography. Annu Int Conf IEEE Eng Med Biol Soc 2021;2021:3916-9. [Crossref] [PubMed]
- Mansoory MS, Ahmadian A, Gorgian Mohammadi A, et al. Mitral valve prolapse detection using landmark extraction from echocardiography sequences. Annu Int Conf IEEE Eng Med Biol Soc 2012;2012:4303-6. [Crossref] [PubMed]
- Bosch JG, Mitchell SC, Lelieveldt BP, et al. Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE Trans Med Imaging 2002;21:1374-83. [Crossref] [PubMed]
- Zych-Krekora K, Sylwestrzak O, Grzesiak M, et al. Impact of Prenatal and Postnatal Diagnosis on Parents: Psychosocial and Economic Aspects Related to Congenital Heart Defects in Children. J Clin Med 2023;12:5773. [Crossref] [PubMed]
- Vena F, Manganaro L, D'Ambrosio V, et al. Neuroimaging and Cerebrovascular Changes in Fetuses with Complex Congenital Heart Disease. J Clin Med 2022;11:6740. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham; 2015:234-41.
- Fan T, Wang G, Li Y. MA-Net: A Multi-Scale Attention Network for Liver and Tumor Segmentation. IEEE Access 2020;8:179656-65.
- Chaurasia A, Culurciello E. LinkNet: Exploiting encoder representations for efficient semantic segmentation. 2017 IEEE Visual Communications and Image Processing (VCIP); 10-13 December 2017; St. Petersburg, FL, USA. IEEE; 2017:1-4.
- Donofrio MT, Moon-Grady AJ, Hornberger LK, et al. Diagnosis and treatment of fetal cardiac disease: a scientific statement from the American Heart Association. Circulation 2014;129:2183-242. [Crossref] [PubMed]
- Digilio MC, Pugnaloni F, De Luca A, et al. Atrioventricular canal defect and genetic syndromes: The unifying role of sonic hedgehog. Clin Genet 2019;95:268-76. [Crossref] [PubMed]
- Miller A, Siffel C, Lu C, et al. Long-term survival of infants with atrioventricular septal defects. J Pediatr 2010;156:994-1000. [Crossref] [PubMed]
- Taqatqa AS, Vettukattil JJ. Atrioventricular Septal Defects: Pathology, Imaging, and Treatment Options. Curr Cardiol Rep 2021;23:93. [Crossref] [PubMed]
- Aly S, Qattea I, Othman H, et al. Outcomes of atrioventricular septal defects with and without down syndrome: analysis of the national inpatient database. Cardiol Young 2023; Epub ahead of print. [Crossref]
- Schumacher K, Marin Cuartas M, Meier S, et al. Long-term results following atrioventricular septal defect repair. J Cardiothorac Surg 2023;18:250. [Crossref] [PubMed]
- Jonas RA. LV outflow obstruction after repair of atrioventricular septal defect: an uncommon but challenging problem. Interact Cardiovasc Thorac Surg 2022;34:611-2. [Crossref] [PubMed]
- Veronese P, Guariento A, Cattapan C, et al. Prenatal Diagnosis and Fetopsy Validation of Complete Atrioventricular Septal Defects Using the Fetal Intelligent Navigation Echocardiography Method. Diagnostics (Basel) 2023;13:456. [Crossref] [PubMed]
- Lockhart MM, Phelps AL, van den Hoff MJ, et al. The Epicardium and the Development of the Atrioventricular Junction in the Murine Heart. J Dev Biol 2014;2:1-17. [Crossref] [PubMed]
- Wessels A, van den Hoff MJ, Adamo RF, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol 2012;366:111-24. [Crossref] [PubMed]
- Rigby M. Atrioventricular Septal Defect: What Is in a Name? J Cardiovasc Dev Dis 2021;8:19. [Crossref] [PubMed]


