
A ferroptosis-related ceRNA network for investigating the molecular mechanisms and the treatment of neonatal hypoxic-ischemic encephalopathy
Highlight box
Key findings
• We have established a competing endogenous RNA (ceRNA) network.
• We have identified three key genes.
• We have predicted four drugs associated with ferroptosis in hypoxic-ischemic brain damage (HIBD).
What is known and what is new?
• There are studies on iron death in HIBD, but there are few studies on the mechanism of iron death.
• This manuscript mainly includes identifying key apoptosis-related genes of HIBD, constructing a lncRNA-miRNA-mRNA network, and further exploring the pathogenesis of HIBD.
What is the implication, and what should change now?
• This paper promotes the further mechanism research of HIBD. We further explored the potential molecular mechanisms of key genes that affect the progression of HIBD, bringing some new ideas for the diagnosis and treatment of HIBD.
• A large number of lncRNAs and miRNAs have been identified in this study, and their expression in HIBD and the established ceRNA network need to be further verified.
Introduction
Neonatal hypoxic-ischemic brain damage (HIBD) represents hypoxic-ischemic brain injury in newborns due to perinatal asphyxia and is one of the most common causes of neurological disease and death in newborns, occurring in 2–3 per 1,000 births (1,2). Unfortunately, specific and effective treatments for this disease have not yet been developed (3). Despite the timely adoption of mild hypothermia treatment, a number of newborns with severe asphyxia may have adverse outcomes, such as cerebral palsy, intellectual disability, epilepsy, and even death. Moreover, adverse outcomes lead to a considerable socio-economic burden. Hence, it is crucial to discover an effective method for the treatment of HIBD (2,4).
The pathophysiological mechanism of HIBD has not fully been deciphered. According to recent data, the development of HIBD can be promoted by various factors, such as disrupted metabolism and a protracted inflammatory response (5). These factors can lead to different types of cell death, a complex process closely related to the development and maintenance of body homeostasis and closely linked to the occurrence, development, and prognosis of various diseases (6). Iron death, also known as ferroptosis, is a unique form of programmed cell death (PCD) characterized by iron accumulation and oxidative stress with distinct morphological, biochemical, and genetic features compared to other types of PCDs such as apoptosis, necrosis, and autophagy (7). The morphology changes of ferroptotic cells mainly include mitochondrial atrophy, increased membrane density, and reduction or disappearance of mitochondrial ridge (7-10). The most relevant biochemical characteristics include increased reactive oxygen species (ROS), decreased intracellular glutathione peroxidase 4, accumulation of lipid peroxides, and perturbations of iron metabolism (9). In the fight against human diseases, including HIBD, many types of immune cells, such as neutrophils, T and B cells, macrophages, natural killer (NK) cells, and dendritic cells, operate as key factors. The function of immune cells is regulated by cell death. Some researchers have found that ferroptosis is essential for regulating the functions and numbers of immune cells, whereas ferroptosis in non-immune cells may induce the release of damage-associated molecular patterns (DAMPs), triggering an immune cell response (11). However, the function of ferroptosis in the pathogenesis of HIBD is unclear. Therefore, exploring the potential role and mechanism of ferroptosis may provide a new strategy and approach to preventing and treating such patients.
The non-coding RNA (ncRNA) is a non-protein-coding transcript containing hidden regulatory signals for gene expression and disease progression. Based on the length, the ncRNAs can be arbitrarily defined as either long non-coding RNAs (lncRNAs, at least 200 nucleotides) or smaller non-coding RNAs such as transfer RNA, microRNA (miRNA) and piwi interacting RNAs (12-14). MiRNAs and lncRNAs participate in many immune response processes based on the modulation of the immune response genes (15). The hypothesis of competing endogenous RNA (ceRNA) revealed a new mechanism of RNA interaction. LncRNA may compete for miRNA-messenger RNA (miRNA-mRNA) binding, thus affecting gene expression mediated by miRNA (16). However, little is known about the expression patterns and the interaction network of ceRNA in HIBD.
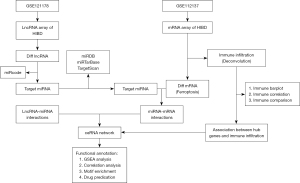
In the present study, we performed a bioinformatic-based analysis of the microarray data of HIBD samples from the Gene Expression Omnibus (GEO). We identified many differentially expressed lncRNAs, miRNAs, and mRNAs, which resulted in establishing a ceRNA network associated with ferroptosis in HIBD. Moreover, our analysis identified three key genes that may play a pivotal regulatory role in modulating ferroptosis and immune cell infiltrations via multiple signaling pathways in this disease. In addition, the drug prediction analysis indicated that four drugs might have a role in ameliorating HIBD-related brain injuries. The ceRNA network and key genes identified in our study might shed new light on the studies of molecular mechanisms and new drug development for HIBD. We present this article in accordance with the ARRIVE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-596/rc).
Methods
Data download
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Data were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/info/datasets.html). The series matrix file of GSE121178 was obtained from the GEO database, and the annotation platform was GPL22120, which contained six samples, including a control group (n=3) and a case group (n=3). The series matrix file of GSE112137 was obtained from the GEO database, and the annotation basis was GPL20301, which contained 16 samples, including a control group (n=8) and a case group (n=8). The 431 ferroptosis-related genes (FRGs) in this study were obtained from the FerrDb V2 database.
Differential expression analysis
Limma is an R package for analyzing differentially expressed genes (DEGs) to identify genes that exhibit obvious differential expression between cases and controls. Data on the molecular mechanisms of neonatal ischemic hypoxic encephalopathy were analyzed by using limma, and DEGs were identified between the two sample groups. The DEGs screening conditions were |log fold change (FC)| >1 and P<0.05. A differential gene volcano map and heat map were drawn.
Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
In order to determine biological functions and pathways involved in disease development, Metascape database (https://metascape.org/) was applied for annotation and visualization; GO analysis and KEGG pathway analysis were conducted for specific genes.
Construction of the lncRNA-miRNA-mRNA network
Recently, ceRNA has attracted much academic attention. It represents a new mode of gene expression regulation. The ceRNA regulatory network is more refined, involving abundant RNA molecules; it includes mRNA, pseudogene encoding genes, miRNA, and lncRNA, among others. miRcode (http://www.mircode.org/) is a popular database for querying lncRNA and miRNA relationships. It covers lncRNA genes with a length of more than 10,000. In this study, we used the miRcode database to predict lncRNA-miRNA interaction pairs. Moreover, three databases, miRDB (http://www.mirdb.org/), miRTarBase (https://mirtarbase.cuhk.edu.cn/), and TargetScan (https://www.targetscan.org/vert_80/), were combined to analyze the interaction of the two kinds RNA. Targeted mRNAs identified by two of the three databases were selected for subsequent research. Then, lncRNA-miRNA interaction and mRNA-miRNA interaction were combined to establish a lncRNA-miRNA-mRNA network, which was visualized by Cytoscape (https://cytoscape.org/).
Analysis of immune cell infiltration
To assess the impact of genes on immune infiltration, the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) method was applied to determine immune infiltration level, and the corrplot package in R was used to analyze the interaction association in immune cells and analyzed the influence of the interaction association. The vioplot package was used to plot the relative content of immune cells, and Pearson analysis was performed for gene expression and immune cell content.
Gene set enrichment analysis (GSEA)
The GSEA method was applied to rank genes based on how differently they were expressed in two groups, and we then checked if the predefined gene sets were enriched according to the list. In this study, by comparing the differences in signaling pathways between the GSEA group and the low-expression group, the molecular mechanism of core genes related to such disease was discussed. The number of replacements was set to 1,000.
Transcriptional regulation analysis of key genes
In the present research, the R package “RcisTarget” was applied to predict transcription factors (TFs) based on motif. The normalized enrichment score (NES) for motifs depends on the total motifs. Except for the motifs annotated by the source data, we inferred further annotated files. The first step in estimating the overexpression of each motif was to calculate the area under the curve (AUC) of every motif-motif set pairs. This was calculated by the recovery curve of the sequence based on the gene set. The motif NES was determined according to the AUC distribution.
Connectivity Map (CMap) drug prediction
The CMap is a gene expression profile database based on intervention gene expression developed by the Broad Institute. It is mainly used to reveal functional associations of small molecule compounds, genes, and disease states. It contains gene chip data before and after 1,309 small molecule drugs are treated with five human cell lines. Treatment conditions are varied, including different drugs, different concentrations, different treatment duration, and so on. In this study, the differentially expressed genes of the disease were used to predict the targeted therapeutic drugs for the disease. The differentially up-regulated and down-regulated mRNAs were uploaded to the CMap database, and L1000 was selected as the background set to predict the potential therapeutic drugs for the disease.
Neonatal HIBD in an animal model
Our model references the Rice-Vannucci rat model (17). A protocol was prepared before the study without registration. All animal experiment procedures were approved by the Ethics Committee of the Nantong University (approval No. S20220321-008) and were conducted according to the institutional animal care and use guidelines for the care and use of animals. The newborn rats were fed freely by their mothers. The conditions were maintained with a room temperature of 24±2 ℃ and a 12-hour light/dark cycle. Rats have free access to food and water. Both male and female pups received the HIBD model. Neonatal Sprague-Dawley (SD) rats (12–17 g, 7 days old) were purchased from the Experimental Center of Nantong University. Six newborn rats were randomly divided into the HIBD group and sham operation control group, with 3 rats in the HIBD group and three rats in the sham operation control group. In this study, a blind method was used to take pup brains and do polymerase chain reaction (PCR). The 7-day-old SD rats in the HIBD group were anesthetized with isoflurane, then the rats were fixed in supine position, the left common carotid artery was exposed, separated from nerves and veins, ligation with 4-0 surgical line, and finally sutured to allow the animals to recover. After surgery, the rats were placed back with their mothers for 1 hour and then placed in a closed anoxic chamber (8% oxygen, 92% nitrogen, flow rate of 4 L/min) on a heating mat at 37 ℃ for 2.5 hours. After hypoxia, the surviving pups were returned to their mothers (there were no dead pups in the experiment). The same procedure was performed in the sham operation control group except that the exposed carotid artery was not lapped. After 24 hours, the rats were decapitated and their brains removed for subsequent tests.
Quantitative PCR
The total RNA of rat brain tissues was prepared by TRIzol, and reverse transcription was performed using a reverse transcription kit. The primers used in quantitative real-time PCR (qRT-PCR) test were as follows: HMOX1, F: 5'-GCCTCTTCTGTCACCCTGT-3', R: 5'-TCTGCTTGGCTTCCTCCC-3'; MYCN, F: 5'-ACCCAACATCAGCGGTCG-3', R: 5'-CGTGACTGTCGGGTT TTCCA-3'; QSOX1, F: 5'-AGCCACTGCCCTAGATGTACC-3', R: 5'-TGAGGCCTGCGTTTAGTTCC-3'. The initial denaturation period of the reaction was 95 ℃ for 10 minutes, then 95 ℃ for 15 seconds, 60 ℃ for 60 seconds, and 95 ℃ for 15 seconds, for a total of 40 cycles. Fluorescence was recorded at each annealing step. After each PCR run, the system automatically analyzed the data and obtained the amplification map. The expression levels of these genes were normalized to endogenous glyceraldehyde 3-phosphate dehydrogenase complementary DNA. The results of the experiment were represented by relative quantitative analysis of 2−△△CT, and all experiments were repeated three times.
Statistical analysis
Most of the statistical analyses in this study were performed using R software (version 4.2.1; The R Foundation for Statistical Computing, Vienna, Austria). The Kolmogorov-Smirnov test was applied to data obtained by the qRT-PCR analysis of the mRNA expression in brain tissues of HIBD and control rats. P<0.05 was considered statistically significant.
Results
Differential expression analysis of lncRNA associated with HIBD and prediction of miRNA
Figure 1 depicts the workflow of the current study. To identify lncRNAs inducing a regulatory function in the HIBD process, we downloaded the GSE121178 dataset from the GEO database and analyzed the DEGs of 6 blood samples, including a control group (n=3) and case group (n=3). We used the limma package to calculate differential lncRNAs in the dataset. The differential gene judgment criteria were P<0.05 and |LogFC| >1. Up to 1,178 differential lncRNAs were screened, including 473 up-regulated lncRNAs and 705 down-regulated lncRNAs (Figure 2A,2B). Subsequently, we used the miRcode database to predict the miRNA targets of the 1,178 differentially expressed lncRNAs. The analysis detected 207 lncRNA-related target miRNAs, which were visualized using Cytoscape (Figure 2C).
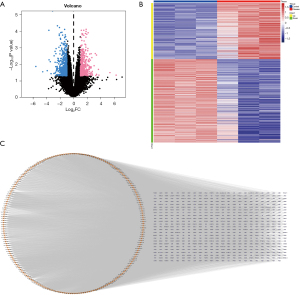
Differentially expressed ferroptosis-related mRNAs in HIBD
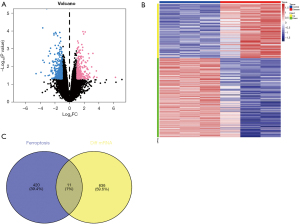
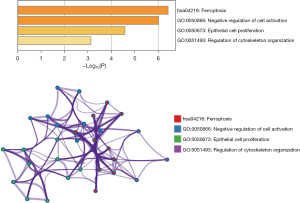
To investigate if the alterations in the expression of lncRNAs and miRNAs affect ferroptosis in HIBD, we screened the differential mRNAs in the GSE112137 dataset downloaded from the GEO database using the limma package. The differential gene judgment criteria were P<0.05 and |LogFC| >1. Up to 647 differential mRNAs were screened, including 264 up-regulated and 383 down-regulated genes (Figure 3A,3B). Subsequently, we intersected these 647 differential mRNAs with FRGs and found 11 intersection genes (Figure 3C). We further conducted a GSEA for the 11 differential ferroptosis mRNAs via Metascape, which indicated that the main enrichment pathways of the differential iron death-related genes include ferroptosis, negative regulation of cell activation, epithelial cell proliferation, and regulation of cytoskeleton organization (Figure 4). These data indicated that the 11 FRGs might be involved in the development of HIBD, possibly by regulating ferroptosis via the enriched pathways.
Establishment of ceRNA network associated with ferroptosis and identification of key genes
We used the miRwalk, miRDB, and miRTarBase databases to predict the target mRNAs of the 207 miRNAs identified above and identified 8,612 mRNAs in the GSE121178 dataset. Finally, the intersection of 8,612 predicted mRNAs and 11 differential FRGs yielded four mRNAs (Figure 5A). The ceRNA network was successfully constructed and visualized via Cytoscape (Figure 5B). In addition, we downloaded the GSE112137 dataset from the GEO database as an external dataset for subsequent analysis. The results indicated that three mRNAs, including HMOX1, MYCN, and QSOX1, were differentially expressed in HIBD samples compared to the normal controls (Figure 5C). Therefore, HMOX1, MYCN, and QSOX1 were selected as the key genes for subsequent analysis.
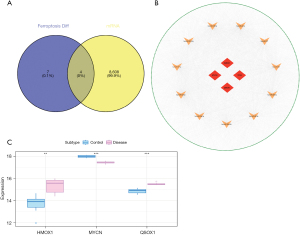
Validation of key genes in an animal model
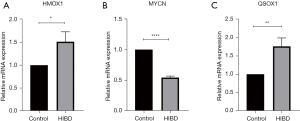
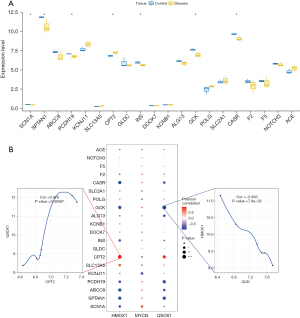
To further verify the expression levels of the three key genes in HIBD, we constructed an animal model of the disease and collected brain tissue samples from the HIBD rats 24 hours after hypoxia treatment. The mRNA levels of the three genes (HMOX1, MYCN, and QSOX1) in these samples were tested using qRT-PCR and compared with those in the brain tissues of the control rats. The results showed that the levels of HMOX1 and QSOX1 in HIBD rats were significantly higher than those in control rats, whereas the expression of MYCN in HIBD rats was significantly reduced compared to that in controls. These results were consistent with findings obtained in the above bioinformatic analysis (Figure 6A-6C).
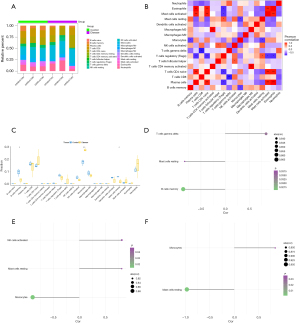
The three key genes correlate with the infiltration of multiple immune cells
The immune microenvironment is mainly composed of immune cells, extracellular matrix (ECM), growth factors, and inflammatory factors, which affect the prognosis and the sensitivity of the disease therapy. Based on the association of key genes and immune infiltration, we further explored key genes’ potential molecular mechanisms in affecting the progression of neonatal hypoxic encephalopathy. The distribution of the infiltrating immune cells and their correlations are shown in Figure 7A,7B. Significant differences were observed regarding the infiltration of the memory B cells, monocytes, and resting mast cells between the disease and control groups (Figure 7C). Subsequently, the association of key genes and immune cells was analyzed, and it was found that the three key genes were closely related to several immune cells. HMOX1 was significantly positively related to gamma delta T cells and negatively related to resting mast cells and memory B cells (Figure 7D). MYCN was positively related to activated NK cells and resting mast cells and negatively correlated with monocytes (Figure 7E). QSOX1 was positively correlated with monocytes and negatively related to resting mast cells (Figure 7F). In addition, we obtained the association between these key genes and immune factors from the TISIDB (http://cis.hku.hk/TISIDB/), including immunosuppressors, immunostimulators, and receptors (Figure 8). These analyses suggest that key genes are significantly associated with the level of immune cell infiltration and play a key role in the immune environment in HIBD.
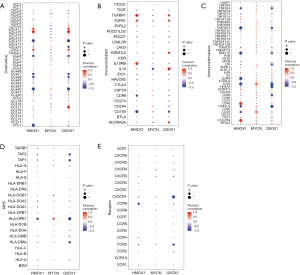
The key genes regulate multiple signaling pathways
The three key genes may be involved in the development of HIBD by regulating some signaling pathways. Thus, we performed GSEA to analyze the specific signaling pathways that may be regulated by HMOX1, MYCN, and QSOX1 and explored the potential molecular mechanisms of the key genes affecting the progression of HIBD. The results of GSEA showed that HMOX1 is enriched in several pathways, such as the NOD-like receptor signaling pathway and oxidative phosphorylation (Figure 9A). MYCN could enrich alanine aspartate and glutamate metabolism, cell adhesion molecules, and other signaling pathways (Figure 9B). QSOX1 enriched several signaling pathways, such as type II diabetes mellitus and ubiquitin-mediated proteolysis (Figure 9C). These data suggest that the three key genes may influence disease progression through multiple signaling pathways.
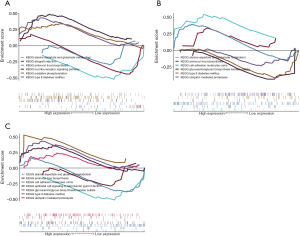
Analysis of transcriptional regulation of the key genes
We applied these three key genes to the gene set used in this study; we found that they are regulated by common mechanisms such as multiple TFs. Therefore, the TFs involved in the modulation of the three key genes were enriched by cumulative recovery curves. The results of motif-TF annotation indicated that the motif with the highest standardized enrichment score (NES: 4.88) was cisbp_M0451. All the enriched motifs and relevant TF results are displayed in Figure 10.
Analysis of the correlation between key genes and disease-regulating genes
To investigate the possible relationship between the key genes and the HBID-regulating genes, we obtained the disease-regulating genes from the GeneCards database (https://www.genecards.org/). We then analyzed the expression levels of the top 20 genes with the highest relevance score in different groups and found that the expressions of SCN1A, PCDH19, CPT2, INS, GCK, CASR, and SCTAN1 were different between cases and controls (Figure 11A). As a result, it can be concluded that the expression levels of the three essential genes were closely related to those of several disease-related genes (Figure 11B). QSOX1 and CPT2 were positively correlated with significance (r=0.976), whereas a significant negative correlation between HMOX1 and GCK was observed (r=−0.993). These data demonstrated that the key genes had significant correlations to HBID-regulating genes.
Potential drug prediction
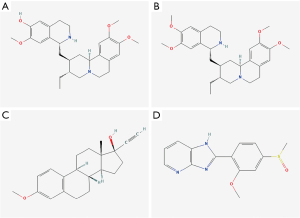
Based on the above analysis, we attempted to predict the effectiveness of drugs for HBID treatment. The top 150 up-regulated mRNAs and the top 150 down-regulated mRNAs in HBID were used for drug prediction through the CMap database. The results showed that perturbations of gene expression induced by the drugs, including cephaeline, mestranol, and sulmazole, were inversely related mainly to those triggered by the disease (Figure 12), suggesting that these drugs could alleviate the disease.
Discussion
The current treatment of HIBD is poorly effective, and great efforts are needed to improve its efficacy for this significant threat to newborn life (18). The current state of the art is mainly due to the unclear and incomplete understanding of the disease pathogenesis. A recent study has revealed that the development of HIBD is associated with ferroptosis, a unique form of cell death depending on iron and lipotoxicity (19). Studies have also shown that inhibiting ferroptosis can improve HIBD in neonates (20,21). It has been reported that ferroptosis is regulated by many cellular metabolic events, such as REDOX homeostasis, iron processing, mitochondrial activity, metabolism of lipids, sugars, and many disease-associated pathways (22). Increasing evidence demonstrates that ferroptosis is associated with cell damage in a variety of diseases, such as cancer, ischemia-reperfusion injury, stroke, neurodegeneration, myocardial infarction, acute kidney injury, and liver injury (23-26). However, the mechanism of ferroptosis in HIBD has not been deeply studied.
Our research seeks to identify ferroptosis-related biomarkers affecting neonatal HIBD, establish ceRNA networks, and explore their roles in HIBD immune cell infiltration. We screened 647 differentially expressed mRNAs through the GSE112137 dataset intersected them with FRGs, and identified 11 DEGs. Enrichment analysis showed that the main enrichment pathways of these 11 genes were ferroptosis, negative regulation of cell activation, cytoskeletal tissue regulation, and epithelial cell proliferation. Moreover, we successfully constructed the ceRNA network through the GSE121178 dataset and intermixed the predicted mRNA with 11 differential ferroptosis genes to obtain four mRNAs. Through external verification, three key genes showed significant differences. Compared with the control group, the expression of HMOX1 and QSOX1 was higher in the HIBD group, whereas the expression of MYCN in the HIBD group was lower than that in the control group. We constructed an animal model of HIBD in which these three key molecules were validated using qRT-PCR, and the results were consistent with those of microarray analysis.
HMOX1, also known as heat shock protein 32, is an enzyme involved in the catabolism of heme induced by various pro-oxidation and inflammatory stimuli (27). It is protective in anti-oxidation, anti-apoptosis, anti-inflammation, and vascular relaxation (28). The expression of HMOX1 contributes to mediating the resolution of inflammation, including neuroinflammation (29). In our study, the enrichment analysis revealed that HMOX1 may be involved in various signaling pathways that influence the development of HIBD, such as the NOD-like receptor signaling pathway and oxidative phosphorylation. Notably, the most studied NOD-like receptor, NLRP3, has significantly higher activity in hypoxic-ischemic encephalopathy (HIE) rats (30). Using drugs to inhibit the inflammasome activity can significantly inhibit HIE-related brain damage (31). Thus, whether HMOX1 can affect HIBD via regulating NOD-like receptors such as NLPR3 deserves further study.
MYCN is a TF of the MYC proto-oncogene family. It is involved in the basic process of controlling embryonic development (32). MYCN promotes cell proliferation, growth, and apoptosis (33-35). A previous study has shown that MYCN is over-expressed in various pediatric cancers, including neuroblastoma, rhabdomyosarcoma, medulloblastoma, nephroblastoma, and retinoblastoma (36). However, until now, no study has described the relationship between this gene and HIBD. QSOX1 is an enzyme that oxidizes mercaptan during protein folding, reducing molecular oxygen to hydrogen peroxide (37). Similarly, no study has shown its role in HIBD. The roles and relative contributions of MYCN and QSOX1 in HIBD need further clarification in future studies. Our study also analyzed the upstream regulation and found that HMOX1, MYCN, and QSOX1 are regulated by common mechanisms such as multiple TFs. Hence, we conducted an enrichment analysis to research these TFs and found that GFI1 is the primary regulator of these three molecules.
Accumulating evidence shows that immune cells are closely related to the pathogenesis of HIBD (38). By using the CIBERSORT, we identified 22 immune cell types infiltrating the brains of HIBD patients. The proportion of monocytes in the HIBD group was high, whereas that of memory B cells and resting mast cells was low. Monocytes belong to the mononuclear phagocyte system and are involved in the immune responses following tissue injury and infection. A study has shown that monocytes promote acute inflammatory response, and monocyte infiltration has significant plasticity, pathological function, and lasting harmful effects in neonatal brain injury (39). At the same time, we verified the association between the three core genes (HMOX1, MYCN, QSOX1) and immune cell infiltration. Our results show that these three core genes correlate with immune cell infiltration and are closely related to gamma delta T cells, resting mast cells, memory B cells, and activated NK cells. These results further demonstrate the key function of the core genes and immune infiltration in the HIBD. Moreover, GSEA also revealed the specific signaling pathways of three core genes that may be involved in the HIBD process; this finding may be considered highly beneficial for a deeper understanding of the pathogenesis of HIBD.
This study identified regulatory genes associated with HIBD through the Genecard database. Seven genes (SCN1A, PCDH19, CPT2, INS, GCK, CASR, and SCTAN1) differed between the two groups. CASR, a G-protein-coupled receptor cloned initially from the bovine parathyroid gland, maintains calcium homeostasis by regulating the secretion of parathyroid hormone (40,41). The results of Pak et al. showed that selective and persistent astrocyte-induced CaSR expression is a common feature of ischemic injury, suggesting a role of CaSR in ischemia-induced astrocyte response (42). We analyzed the correlation between three core genes and HIBD regulatory genes and revealed the close relationships between the genes. The mechanism of the three core genes and HIBD regulatory genes in HIBD is still unclear and needs further exploration.
The CMap database was used for drug prediction, and the results showed that the expression profiles of drug disturbances in cephaeline, emetine, mestranol, and sulmazole were negatively related to the expression result of disease disturbances, suggesting that these drugs could alleviate the disease state. These findings provide new boosts for developing innovative, more effective drugs to treat HIBD.
As described above, our study provides new insights for further investigation of the molecular mechanisms of HIBD, even though the study has some limitations. Firstly, although we identified numerous lncRNAs and miRNAs, their expression and the established ceRNA network in HIBD should be confirmed by further studies. Secondly, there is a lack of data about the related clinical features and prognostic data in HIBD patients; clinical relevance studies, prognostic analyses of the ceRNA network, and the key genes should be conducted in future studies. Thirdly, the number of patients in the database used in this study is relatively small, and thus, the results need to be further validated in a study involving a larger sample of HIBD patients. Despite these limitations, our findings contribute to elucidating the function and mechanism of ferroptosis in HIBD.
Conclusions
In this study, after screening out 11 differential FRGs in HIBD, bioinformatics analysis identified three key genes associated with ferroptosis and the development of HIBD, including HMOX1, MYCN, and QSOX1. The study also established a ceRNA network in HIBD, which might promote further mechanistic studies of this disease. The potential molecular mechanism of key genes affecting the progression of HIBD was further explored and yielded some new ideas regarding the diagnosis and treatment of HIBD.
Acknowledgments
Funding: This work was supported partially by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-596/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-596/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-596/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-596/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All animal experiment procedures were approved by the Ethics Committee of the Nantong University (approval No. S20220321-008) and were conducted according to the institutional animal care and use guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Finder M, Boylan GB, Twomey D, et al. Two-Year Neurodevelopmental Outcomes After Mild Hypoxic Ischemic Encephalopathy in the Era of Therapeutic Hypothermia. JAMA Pediatr 2020;174:48-55. [Crossref] [PubMed]
- Disdier C, Stonestreet BS. Hypoxic-ischemic-related cerebrovascular changes and potential therapeutic strategies in the neonatal brain. J Neurosci Res 2020;98:1468-84. [Crossref] [PubMed]
- Lee BL, Glass HC. Cognitive outcomes in late childhood and adolescence of neonatal hypoxic-ischemic encephalopathy. Clin Exp Pediatr 2021;64:608-18. [Crossref] [PubMed]
- Juul SE, Comstock BA, Heagerty PJ, et al. High-Dose Erythropoietin for Asphyxia and Encephalopathy (HEAL): A Randomized Controlled Trial - Background, Aims, and Study Protocol. Neonatology 2018;113:331-8. [Crossref] [PubMed]
- Lin W, Zhang T, Zheng J, et al. Ferroptosis is Involved in Hypoxic-ischemic Brain Damage in Neonatal Rats. Neuroscience 2022;487:131-42. [Crossref] [PubMed]
- Wolpaw AJ, Shimada K, Skouta R, et al. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc Natl Acad Sci U S A 2011;108:E771-80. [Crossref] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. [Crossref] [PubMed]
- Yagoda N, von Rechenberg M, Zaganjor E, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007;447:864-8. [Crossref] [PubMed]
- Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis 2020;11:88. [Crossref] [PubMed]
- Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol 2008;15:234-45. [Crossref] [PubMed]
- Chen X, Kang R, Kroemer G, et al. Ferroptosis in infection, inflammation, and immunity. J Exp Med 2021;218:e20210518. [Crossref] [PubMed]
- Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol 2017;35:177-98. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26-46. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Manco G, Lacerra G, Porzio E, et al. ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives. Biomolecules 2022;12:443. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Rice JE 3rd, Vannucci RC, Brierley JB. The influence of immaturity on hypoxic-ischemic brain damage in the rat. Ann Neurol 1981;9:131-41. [Crossref] [PubMed]
- Cai Y, Li X, Tan X, et al. Vitamin D suppresses ferroptosis and protects against neonatal hypoxic-ischemic encephalopathy by activating the Nrf2/HO-1 pathway. Transl Pediatr 2022;11:1633-44. [Crossref] [PubMed]
- Sun Y, Chen P, Zhai B, et al. The emerging role of ferroptosis in inflammation. Biomed Pharmacother 2020;127:110108. [Crossref] [PubMed]
- Zhang M, Lin W, Tao X, et al. Ginsenoside Rb1 inhibits ferroptosis to ameliorate hypoxic-ischemic brain damage in neonatal rats. Int Immunopharmacol 2023;121:110503. [Crossref] [PubMed]
- Li Y, Wang T, Sun P, et al. Farrerol Alleviates Hypoxic-Ischemic Encephalopathy by Inhibiting Ferroptosis in Neonatal Rats via the Nrf2 Pathway. Physiol Res 2023;72:511-20. [Crossref] [PubMed]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22:266-82. [Crossref] [PubMed]
- Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res 2019;29:347-64. [Crossref] [PubMed]
- Li W, Feng G, Gauthier JM, et al. Ferroptotic cell death and TLR4/Trif signaling initiate neutrophil recruitment after heart transplantation. J Clin Invest 2019;129:2293-304. [Crossref] [PubMed]
- Hou L, Huang R, Sun F, et al. NADPH oxidase regulates paraquat and maneb-induced dopaminergic neurodegeneration through ferroptosis. Toxicology 2019;417:64-73. [Crossref] [PubMed]
- Hu Z, Zhang H, Yi B, et al. VDR activation attenuate cisplatin induced AKI by inhibiting ferroptosis. Cell Death Dis 2020;11:73. [Crossref] [PubMed]
- Sharp FR, Zhan X, Liu DZ. Heat shock proteins in the brain: role of Hsp70, Hsp 27, and HO-1 (Hsp32) and their therapeutic potential. Transl Stroke Res 2013;4:685-92. [Crossref] [PubMed]
- Bereczki D Jr, Balla J, Bereczki D. Heme Oxygenase-1: Clinical Relevance in Ischemic Stroke. Curr Pharm Des 2018;24:2229-35. [Crossref] [PubMed]
- Syapin PJ. Regulation of haeme oxygenase-1 for treatment of neuroinflammation and brain disorders. Br J Pharmacol 2008;155:623-40. [Crossref] [PubMed]
- Wang L, Hauenstein AV. The NLRP3 inflammasome: Mechanism of action, role in disease and therapies. Mol Aspects Med 2020;76:100889. [Crossref] [PubMed]
- Zhu JJ, Yu BY, Huang XK, et al. Neferine Protects against Hypoxic-Ischemic Brain Damage in Neonatal Rats by Suppressing NLRP3-Mediated Inflammasome Activation. Oxid Med Cell Longev 2021;2021:6654954. [Crossref] [PubMed]
- Ruiz-Pérez MV, Henley AB, Arsenian-Henriksson M. The MYCN Protein in Health and Disease. Genes (Basel) 2017;8:113. [Crossref] [PubMed]
- Ham J, Costa C, Sano R, et al. Exploitation of the Apoptosis-Primed State of MYCN-Amplified Neuroblastoma to Develop a Potent and Specific Targeted Therapy Combination. Cancer Cell 2016;29:159-72. [Crossref] [PubMed]
- Chen L, Iraci N, Gherardi S, et al. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res 2010;70:1377-88. [Crossref] [PubMed]
- Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis 2003;8:115-28. [Crossref] [PubMed]
- Liu Z, Chen SS, Clarke S, et al. Targeting MYCN in Pediatric and Adult Cancers. Front Oncol 2020;10:623679. [Crossref] [PubMed]
- Lake DF, Faigel DO. The emerging role of QSOX1 in cancer. Antioxid Redox Signal 2014;21:485-96. [Crossref] [PubMed]
- O'Hare FM, Watson RW, O'Neill A, et al. Persistent systemic monocyte and neutrophil activation in neonatal encephalopathy. J Matern Fetal Neonatal Med 2016;29:309-16. [Crossref] [PubMed]
- Chen HR, Chen CW, Kuo YM, et al. Monocytes promote acute neuroinflammation and become pathological microglia in neonatal hypoxic-ischemic brain injury. Theranostics 2022;12:512-29. [Crossref] [PubMed]
- Brown EM, MacLeod RJ. Extracellular calcium sensing and extracellular calcium signaling. Physiol Rev 2001;81:239-97. [Crossref] [PubMed]
- Brown EM, Gamba G, Riccardi D, et al. Cloning and characterization of an extracellular Ca(2+)-sensing receptor from bovine parathyroid. Nature 1993;366:575-80. [Crossref] [PubMed]
- Pak HJ, Riew TR, Shin YJ, et al. Enhanced expression of the calcium-sensing receptor in reactive astrocytes following ischemic injury in vivo and in vitro. J Neurol Sci 2016;366:102-9. [Crossref] [PubMed]



