
Polymorphic markers of several immune regulatory genes modulate the susceptibility for eczema and related phenotypes in children
Highlight box
Key findings
• Transforming growth factor-beta 1 (TGFB1)_rs1800469 is associated with childhood eczema (including atopic eczema) and allergic rhinitis.
• Interleukin-10 (IL10) may be associated with increased total immunoglobulin E (IgE) levels.
• Individuals’ total IgE levels may be modulated by particular single-nucleotide polymorphisms (SNPs) of immune regulatory genes (IL10_rs3021094, TGFB1_rs1800469, IL-6 receptor_rs2228145, and signal transducer and activator of transcription 3_rs4796793).
What is known and what is new?
• Certain gene mutations are associated with eczema susceptibility in the Caucasian population, but the previously identified eczema susceptibility gene polymorphisms in Caucasians have limited generalizability in Asians due to lower prevalences.
• This study serves to provide data on genetic associations between eczema and its related subphenotypes with certain immune regulatory gene SNPs particularly of the southern Chinese population, which was previously lacking in literature.
What is the implication, and what should change now?
• This genetic association study supports the importance of immune regulatory genes in the pathogenesis of eczema in children.
• Testing for polymorphic markers of TGFB1 and other immune regulatory genes facilitates the risk prediction for childhood eczema.
Introduction
According to the Global Burden of Disease Study in 2019, eczema has an age-standardized prevalence rate of 2.28%, with peak prevalence in children aged between five and nine (1). It accounts for 36.17% of immune-mediated inflammatory disease cases and is more commonly observed in individuals with higher socio-demographic status (1,2). The treatment modality of choice is individualized, primarily depending on patients’ signs and symptoms (3). Damiani et al. established an Italian guideline to maintain a standard of care for eczema control and guide practitioners in adopting the novel biologic—dupilumab for eczema (3).
The pathogenesis of eczema is multifactorial with environmental factors and genetic predisposition; these patients are thought to have skin barrier defects accompanied by skin microbial dysbiosis (4). Studies from the coronavirus disease 2019 (COVID-19) pandemic suggested mask-wearing may be associated with exacerbated eczema (5), possibly owing to the increased abundance of Malassezia fungi on skin (6,7). The composition of cutaneous microbiome not only varies between healthy and eczematous individuals, but also with age among the latter group (8). Genome-wide association study (GWAS) identified complex network of genes for eczema and allergic diseases. Elevated immunoglobulin E (IgE), a common laboratory finding in patients with atopic dermatitis, positively correlated with eczema severity and other allergic symptoms. IgE is produced by plasma cells as part of the adaptive immune response; this pathway is promoted by type 2 helper T (Th2) cell-mediated cytokines interleukin (IL)-4 and IL-13 (9). The abovementioned dupilumab acts against IL-4 receptors, thus hindering receptor-ligand binding and its downstream pathway (10).
Despite recent success of GWAS, previously studied populations only shared a limited number of candidate genes for the association with different eczema phenotypes; few were tested for epistatic interactions along the gene network. The main eczema predisposition genes included in the studies were those encoding epidermal barrier protein filaggrin (FLG) and Th2 pathway (11,12). FLG on chromosome 1q21 was the most studied and replicated gene for eczema in Caucasian children, yet many loss-of-function mutations of FLG that were identified in Europeans are less frequently detected in Asians.
Our group screened five published FLG mutations in southern Chinese children, and all of them were rare in our population (13). Besides genetics, the laboratory findings in eczema patients, such as blood IgE and eosinophil percentage (eos%), were different among eczematous patients of similar disease severity between Asian and Caucasian populations (14). Since corresponding data for the Asian population is lacking, our study serves to fill this knowledge gap.
Transforming growth factor-beta 1 (TGF-β1) interacts with multiple cell types to inhibit cell proliferation and apoptosis (15). IL-10, produced by B1 lymphocytes and found in normal skin (16), serves two main functions: (I) inhibiting synthesis of pro-inflammatory cytokines; and (II) inducing class switching in B cells (17). IL-6 is crucial for skin barrier repair and for forming complexes with IL-6 receptor (IL6R), which then migrate to the damaged epidermal layers for repairing permeability barriers through increased signal transducer and activator of transcription 3 (STAT3) phosphorylation (18). STAT3 is a key transcription regulator with multiple roles in inflammation and immune responses; mutations of this gene are associated with immunological diseases such as hyper-IgE syndrome. STAT3 activation is also involved in IL-10 and IL-6 anti-inflammatory signaling.
This study investigated the genetic associations between eczema and related subphenotypes with single-nucleotide polymorphisms (SNPs) of four major immune regulatory genes TGFB1, IL10, IL6R, and STAT3 in southern Chinese children. We present this article in accordance with the MDAR and STROBE reporting checklists (available at https://tp.amegroups.com/article/view/10.21037/tp-23-474/rc).
Methods
Subjects
This study recruited unrelated children, including those without siblings, with physician-diagnosed eczema and non-allergic controls from both pediatric clinics of our university-affiliated teaching hospital and several community-based studies in Hong Kong (19-22). The latter community-based studies were conducted in local schoolchildren primarily for elucidating the epidemiology of childhood obesity and metabolic syndrome (23-26). The participating subjects who suffered from physician-diagnosed eczema and those free from any allergic disease were also recruited into this study. Their blood samples were subjected to our SNP genotyping and specific IgE (sIgE) measurement. All subjects were aged 18 years or younger, whose parents reported themselves as Chinese. Patients’ eczema severity was evaluated by SCORing Atopic Dermatitis (SCORAD). Controls did not have history of eczema, asthma, and allergic rhinitis. Demographics, early-life events and environmental exposures were recorded by the validated Chinese questionnaire of the International Study of Asthma and Allergies in Childhood. The Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee approved this study (Nos. 2008.123 and 2016.171). Subjects and/or their parents gave informed written consent to participate in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Eczema subphenotypes
The subphenotypes of eczema refer to factors which predict progression to eczema and its severity (27). Eos%, expressed as the percentage of total peripheral blood leukocytes, positively correlates with serum total IgE concentration, early eczema onset, and persistence of eczematous lesions (28). Allergen sensitization, measured by skin prick test (SPT) or IgE levels, is often present in eczematous children (19,20,29,30).
Serum total IgE concentration was measured by microparticle immunoassay (IMx Analyzer; Abbott Laboratories, Abbott Park, IL, USA), then log-transformed (logIgE) before analysis. Peripheral blood eosinophils and other leukocytes were counted by Coulter STKS Counter (Beckman Coulter, Brea, CA, USA). Allergen sensitization was assessed by either SPT with standardized crude extracts of Dermatophagoides pteronyssinus, cat dander, and mixed cockroaches (ALK Abelló, Round Rock, TX, USA) or by plasma allergen-sIgE levels to the same allergens by fluorescent enzyme immunoassay (AutoCAP, Phadia AB, Uppsala, Sweden) as decided by the individual studies. In general, SPT was used in studies involving subjects recruited in hospitals while blood IgE assays were used in community-based studies. SPT results with wheal ≥3 mm larger than negative controls and sIgE ≥0.35 kIU/L were considered as positive. Subjects were defined as “atopic” if they had ≥ one positive SPT or sIgE result.
SNP selection and genotyping
Table 1 describes nine SNPs of the four genes being genotyped in this study. The tagging strategy was applied for SNPs with minor allele frequency ≥0.05 and pairwise r2≥0.8 that were within 5-kb both upstream and downstream from the top eczema-associated SNP in the respective target gene. SNPs published to have significant associations with eczema were all included in SNP selection. Nine SNPs selected (four for IL10, one for TGFB1, three for IL6R, and one for STAT3) were genotyped by TaqMan SNP Genotyping assays (Applied Biosystems, Waltham, MA, USA) using a 12-K Quant Studio thermocycler (Applied Biosystems).
Table 1
| Gene | SNP | Position | Major/minor allele | Genotyping efficiency (%) | HWE in controls | HWE in cases |
|---|---|---|---|---|---|---|
| IL10 | rs1800872 | 1q32.1 | T/G | 98.0 | 0.05 | 0.75 |
| rs1800896 | T/C | 99.1 | 0.32 | 0.65 | ||
| rs3790622 | G/A | 99.2 | 0.27 | 0.76 | ||
| rs3021094 | G/T | 98.6 | 0.66 | 0.43 | ||
| TGFB1 | rs1800469 | 19q13.2 | A/G | 97.6 | 0.52 | 0.29 |
| IL6R | rs2228145 | 1q21.3 | A/C | 97.5 | 0.55 | 0.95 |
| rs6689393 | A/G | 98.9 | 0.36 | 0.80 | ||
| rs4845374 | A/T | 98.4 | 0.51 | 0.99 | ||
| STAT3 | rs4796793 | 17q21.2 | C/G | 98.6 | 0.81 | 0.82 |
SNP, single-nucleotide polymorphism; HWE, Hardy-Weinberg equilibrium; IL10, interleukin-10; TGFB1, transforming growth factor-beta 1; IL6R, IL-6 receptor; STAT3, signal transducer and activator of transcription 3.
Statistical analysis
Allele frequencies of SNPs were estimated by gene counting method, and χ2 or Fisher exact test was used to determine Hardy-Weinberg equilibrium (HWE). Pairwise linkage disequilibrium (LD) coefficient was calculated for each SNP pair by HaploView software (Daly Lab, Boston, MA, USA). Associations between SNPs with dichotomous eczema and allergy outcomes were analyzed by logistic regression and those with eczema subphenotypes by linear regression, adjusting for age and sex as covariates.
Epistatic interactions between SNPs for eczema and its subphenotypes were evaluated using generalized multifactor dimensionality reduction (GMDR) beta version 0.9. (http://ibi.zju.edu.cn/software/GMDR/download.html); GMDR computed the maximum-likelihood estimates and the score values of all individuals under the null hypothesis. In the evaluation of the interactive models, outcome parameters including (I) testing accuracy (TA); (II) cross-validation consistency (CVC); and (III) statistical significance of the model were considered. TA, which measures the ability to classify individuals with respect to their score values, was used to choose the best model; a TA score of 0.5 indicated that the model is equal to 50% chance of correct prediction, whereas a score of 1.0 indicates a perfect prediction. CVC evaluated the consistency with which the selected interaction was classified as the best model among all possible combinations. The statistical significance of a model was determined by comparing the average prediction error from the observed data with the distribution of average prediction errors under the null hypothesis of the absence of association, derived empirically from 5,000 permutations. The null hypothesis was rejected when the P value derived from permutation was <0.05.
Subjects were classified by GMDR into low-risk and high-risk groups; their genotypes predicted the respective risks for having eczematous phenotypes and subphenotypes such as high eos% and raised total IgE levels. The results were stratified by subjects’ eczema status. Ten-fold cross-validation was performed, and the possible number of loci was set to include our nine chosen SNPs, as well as additional FLG SNPs and SNPs on the chromosome 11q13 locus (19,22) that our group previously published for eczema phenotypes. One-way analysis of variance (ANOVA) and Kruskal-Wallis test with post-hoc tests were used to compare logIgE level and eos% among different risk groups classified by GMDR. P<0.006 (0.05/9) was considered statistically significant after Bonferroni correction to adjust for multiple statistical testing.
Results
A total of 1,329 eczematous children and 1,179 non-allergic controls were recruited; data for logIgE level and eos% were available in 1,487 and 1,332 subjects, respectively. Table 2 summarises the clinical features of subjects. Control group subjects were significantly older than those with eczema (mean, 13.6 vs. 11.2 years; P<0.001). Subjects with eczema had significantly higher eos%, logIgE level, and atopy values than controls (P<0.001).
Table 2
| Variable | Eczema | Control | P |
|---|---|---|---|
| Sex, n (%) | n=1,329 | n=1,179 | 0.001 |
| Male | 676 (50.9) | 519 (44.0) | |
| Female | 653 (49.1) | 660 (56.0) | |
| Age (years), mean ± SD | 11.2±4.3 | 13.6±4.5 | <0.001 |
| Eos% | n=725 | n=607 | |
| Mean ± SD | 6.9±4.9 | 2.7±2.4 | <0.001 |
| LogIgE level | n=861 | n=626 | |
| Mean ± SD | 2.7±0.8 | 1.8±0.7 | <0.001 |
| Atopy† | n=946 | n=553 | |
| Mean ± SD | 759 (80.2) | 296 (53.5) | <0.001 |
| SCORAD | n=398 | n=0 | |
| Mean ± SD | 30.8±19.4 | NA | NA |
| With moderate-to-severe eczema (objective SCORAD ≥15) (%) |
57.3 | NA | NA |
| History of allergic rhinitis (%) | 66.3 | NA | NA |
| History of asthma (%) | 31.8 | NA | NA |
†, defined as ≥ one positive result by SPT or allergen-sIgE to house dust mite, cat and cockroach. SD, standard deviation; eos%, eosinophil percentage; logIgE, log-transformed total IgE; IgE, immunoglobulin E; SCORAD, SCORing Atopic Dermatitis; NA, not applicable; sIgE, specific IgE.
All nine SNPs were successfully genotyped in over 97.5% of samples, and they met HWE (Table 1). TGFB1_rs1800469 was associated with both eczema [odds ratio (OR), 0.82; 95% confidence interval (CI): 0.73–0.92; P=0.001] (Table 3) and atopic eczema (OR, 0.83; 95% CI: 0.72–0.95; P=0.009), with the former association still significant after adjusting for coexisting asthma (P=0.005). This SNP was also associated with allergic rhinitis (OR, 0.84; 95% CI: 0.74–0.95; P=0.005) (Table 4). TGFB1_rs1800469 was not associated with other subphenotypes of eczema (Tables S1,S2), while STAT3_rs4796793 was inversely associated with SCORAD for patients’ eczema severity when analysed by the dominant model (β, −8.35; 95% CI: −14.10 to −2.60; P=0.005) (Table S2). The IL6R GT haplotype from rs6689393 and rs4845374 was associated with eczema but not atopic eczema (P=0.047) (Table S3). This association, however, became insignificant after Bonferroni correction.
Table 3
| Gene | SNP | Position | Alleles (major/minor) |
MAF (%) | Eczema | Atopic eczema | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Eczema | OR (95% CI) | P† | OR (95% CI) | P† | ||||||
| IL10 | rs1800872 | 1q32.1 | T/G | 29.8 | 28.9 | 0.95 (0.84–1.08) | 0.409 | 0.94 (0.81–1.09) | 0.380 | ||
| rs1800896 | 1q32.1 | T/C | 4.7 | 4.8 | 1.04 (0.79–1.36) | 0.802 | 1.15 (0.84–1.57) | 0.379 | |||
| rs3790622 | 1q32.1 | G/A | 5.2 | 5.3 | 0.94 (0.72–1.22) | 0.628 | 1.00 (0.74–1.35) | 0.976 | |||
| rs3021094 | 1q32.1 | T/G | 46.9 | 47.6 | 1.02 (0.91–1.14) | 0.750 | 1.02 (0.89–1.16) | 0.814 | |||
| TGFB1 | rs1800469 | 19q13.2 | A/G | 44.3 | 39.6 | 0.82 (0.73–0.92) | 0.001 | 0.83 (0.72–0.95) | 0.009 | ||
| IL6R | rs2228145 | 1q21.3 | A/C | 33.2 | 36.4 | 1.13 (1.00–1.28) | 0.053 | 1.15 (0.99–1.33) | 0.062 | ||
| rs6689393 | 1q21.3 | A/G | 45.4 | 48.2 | 1.10 (0.98–1.23) | 0.124 | 1.05 (0.92–1.21) | 0.456 | |||
| rs4845374 | 1q21.3 | A/T | 12.8 | 11.9 | 0.94 (0.79–1.12) | 0.485 | 0.81 (0.65–1.00) | 0.052 | |||
| STAT3 | rs4796793 | 17q21.2 | C/G | 41.9 | 43.2 | 1.09 (0.97–1.23) | 0.149 | 1.04 (0.91–1.20) | 0.572 | ||
†, adjusted for age and sex as covariates. SNP, single-nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; IL10, interleukin-10; TGFB1, transforming growth factor-beta 1; IL6R, IL-6 receptor; STAT3, signal transducer and activator of transcription 3.
Table 4
| Gene | SNP | MAF (%) | Adjusted OR (95% CI)† |
P | |
|---|---|---|---|---|---|
| Control | Allergic rhinitis | ||||
| IL10 | rs1800872 | 30.4 | 28.0 | 0.87 (0.77–0.99) | 0.041 |
| rs1800896 | 4.5 | 5.3 | 1.11 (0.84–1.47) | 0.448 | |
| rs3790622 | 5.1 | 5.1 | 0.98 (0.75–1.28) | 0.870 | |
| rs3021094 | 47.6 | 46.5 | 0.95 (0.85–1.07) | 0.396 | |
| TGFB1 | rs1800469 | 43.2 | 39.1 | 0.84 (0.74–0.95) | 0.005 |
| IL6R | rs2228145 | 34.1 | 36.0 | 1.07 (0.95–1.22) | 0.277 |
| rs6689393 | 46.4 | 47.5 | 1.03 (0.91–1.16) | 0.613 | |
| rs4845374 | 12.6 | 11.8 | 0.95 (0.79–1.14) | 0.591 | |
| STAT3 | rs4796793 | 42.7 | 42.5 | 1.02 (0.90–1.15) | 0.770 |
†, analysed by additive model and adjusted for age and sex as covariates. SNP, single-nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval; IL10, interleukin-10; TGFB1, transforming growth factor-beta 1; IL6R, IL-6 receptor; STAT3, signal transducer and activator of transcription 3.
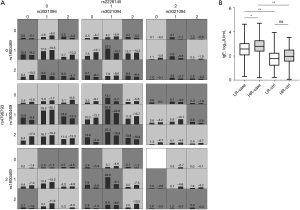
GMDR analysis revealed significant epistatic interaction among four SNPs rs3021094, rs1800469, rs2228145 and rs4796793 for logIgE level after adjustment for age and sex (TA, 0.551; CVC, 10/10; P=0.014) (Table 5). The mean ± standard deviation logIgE levels in high-risk cases, low-risk cases, high-risk controls, and low-risk controls as assigned by GMDR were 2.75±0.79, 2.60±0.80, 1.90±0.66, and 1.81±0.71 respectively (P=0.019 by ANOVA) (Figure 1). The addition of FLG SNPs and 11q13 SNPs did not yield any significant finding for total IgE (Table S4). We did not detect any epistatic interaction for eczema (Table S5), atopic eczema (Table S6), eos%, and SCORAD subphenotypes.
Table 5
| Model formed by IL10, TGFB1, IL6R, and STAT3 | CVC | TA | Cutoff TA 0.05 | Cutoff TA 0.01 | P |
|---|---|---|---|---|---|
| rs2228145 | 9 | 0.523 | 0.534 | 0.543 | 0.149 |
| rs3021094_rs2228145 | 10 | 0.523 | 0.537 | 0.552 | 0.178 |
| rs3021094_rs1800469_rs2228145 | 5 | 0.518 | 0.538 | 0.554 | 0.236 |
| rs3021094_rs1800469_rs2228145_rs4796793† | 10† | 0.551† | 0.538† | 0.554† | 0.014† |
| rs1800872_rs3021094_rs1800469_rs2228145_rs4796793 | 5 | 0.518 | 0.540 | 0.555 | 0.226 |
| rs1800872_rs3021094_rs1800469_rs2228145_rs6689393_rs4796793 | 9 | 0.492 | 0.540 | 0.555 | 0.640 |
| rs1800872_rs1800896_rs3021094_rs1800469_rs2228145_rs6689393_rs4796793 | 8 | 0.530 | 0.539 | 0.558 | 0.113 |
| rs1800872_rs1800896_rs3790622_rs3021094_rs1800469_rs2228145_rs6689393_rs4796793 | 9 | 0.526 | 0.541 | 0.556 | 0.149 |
| rs1800872_rs1800896_rs3790622_rs3021094_rs1800469_rs2228145_rs6689393_rs4845374_rs4796793 | 10 | 0.522 | 0.541 | 0.556 | 0.181 |
†, significant 4-locus model for epistatic interaction. GMDR, generalized multifactor dimensionality reduction; logIgE, log-transformed total IgE; IgE, immunoglobulin E; IL10, interleukin-10; TGFB1, transforming growth factor-beta 1; IL6R, IL-6 receptor; STAT3, signal transducer and activator of transcription 3; CVC, cross-validation consistency; TA, testing accuracy.
Discussion
Immune regulation is mostly mediated by two immunosuppressive cytokines, TGF-β1 and IL-10, that inhibit the activity of both Th1 and Th2 lymphocytes. Similar studies suggested epistatic interactions between immunoregulatory genes may contribute to allergic conditions, such as that among IL2RA, TLR2, TGFΒR2, and FOXP3 for asthma in Europeans (31).
This genetic association study focused on four important immune regulatory genes for eczema and related allergies in 2,508 southern Chinese children. We found the functional SNP rs1800469 of TGFB1 to be associated with both eczema and allergic rhinitis, apart from observing epistatic interaction among IL10_rs3021094, TGFB1_rs1800469, IL6R_rs2228145, and STAT3_rs4796793 for modulating total IgE, which is an important eczema subphenotype (Table 5), through GMDR analysis. In particular, the contribution of IL6R polymorphism in atopic dermatitis aligns with a recently published study involving Asian children (32). These results supported the importance of immune regulatory genes in conferring susceptibility for eczema and related subphenotypes in children. The addition of SNPs of FLG and 11q13 locus (17,22) did not yield additional significant results.
Previous trials investigated any association between TGFB1 and eczema. In a mouse model with eczema, subcutaneous injection of recombinant TGF-β1 suppressed skin lesions and reduced IgE levels (33), suggesting negative correlation between TGF-β1 and eczema severity. Arkwright et al. examined functional SNPs of these two cytokines in children with moderate-to-severe chronic eczema (34). They found that G915C polymorphism of TGFB1, being a low producer genotype, was associated with increased eczema risk.
Our SNP of interest, rs1800469, is located at position -1347 of TGFB1 promoter—two of its alleles were linked to a nearly two-fold difference in plasma TGF-β1 levels, due to competitive binding with activator protein 1 (AP1) and hypoxia-inducible factor 1 (35). The binding of AP1 resulted in transcriptional suppression of TGFB1 and downregulation of other genes. While TGF-β1 itself serves a general immunomodulatory role in preventing allergic diseases, TGFB1 polymorphism and mutations predict increased risks for asthma and increased IgE (36-38).
In our study, TGFB1_rs1800469 was associated with eczema (OR, 0.82; 95% CI: 0.73–0.92), atopic eczema (OR, 0.83; 95% CI: 0.72–0.95) and allergic rhinitis (OR, 0.84; 95% CI: 0.74–0.95), with statistical significance. These observations supported the postulation that TGFB1 polymorphism was associated with allergy outcomes.
According to a previous study, IL-10 was associated with asthma and serum IgE levels (39). This study, however, did not detect significant association between IL10 and childhood eczema by single SNP and haplotype analyses although we identified a trend towards an association with total IgE levels (i.e., did not pass Bonferroni correction). Our finding of low minor allele frequency (0.05) of IL10_rs1800896 (-1082 A/G) was consistent with a Chinese report which found low frequency of 0.29% for GG genotype of this SNP (40). The frequencies of minor alleles for rs1800896 and rs3790622 of IL10 as well as its AG haplotype were around 5%. At this allele frequency, our sample size of 1,329 cases and 1,179 controls had 84% power to detect OR 3 for any association between IL10 allele and eczema with 95% confidence, which would dramatically decrease to 26% power to detect OR of 2 for eczema. Future studies must recruit sufficient sample size to examine the genetic associations for IL10.
Single SNPs of IL6R were not associated with eczema or allergic rhinitis in this study, whereas the GT haplotype from rs6689393 and rs4845374 was marginally associated with eczema but did not survive Bonferroni correction (Table S3). Similar IL6R haplotypes have been reported in other complex diseases such as rheumatoid arthritis and coronary heart disease (41,42). One of our studied IL6R SNPs (rs2228145; Asp358Ala) was associated with persistent eczema, where carriers of the risk allele have increased serum levels of soluble IL-6R (43). The IL-6 cytokine system is involved in skin barrier repair (44), where IL-6 and IL-6R complex migrate to damaged epidermal layers and promote permeability repair through increased STAT3 phosphorylation.
Interestingly, our single SNP analysis revealed significant association between STAT3_rs4796793 and eczema severity as measured by SCORAD (Table S2). This STAT3 SNP was also associated with eczema in Japanese (45).
This study was limited by the lack of functional data of our SNPs and haplotypes to delineate the possible mechanisms that linked different immune regulatory genes. Besides, the eczema subphenotypes of total IgE levels, eos%, and SCORAD were available or recorded only in a subgroup of recruited patients (Table 2). For example, atopy could only be assessed in 946 cases and 553 controls in total. Among the latter, 97 controls were detected by SPT and 456 controls were identified by allergen-sIgE assays. This study found high rate of atopy among the controls who were free from any asthma, rhinitis, and eczema. Although this atopy prevalence was high in our controls, this finding was consistent with our earlier genetic studies of Hong Kong children with different allergic diseases (19,20,22,46,47). The reasons accounting for this high rate of atopy are unclear, but such may be related to high indoor exposure to house dust mites and other inhalant allergens (48,49). Besides, our controls were significantly older (difference in mean of 2.4 years) than the cases (Table 2), suggesting that their clinical status as controls—freed from allergic diseases—was more reliable at an older age. This study was also limited to examining the SNPs of TGFB1, IL10, IL6R, and STAT3. Future research may investigate the associations between SNPs of the IL4 gene and variations in efficacy and adverse drug reactions of dupilumab efficacy (50) as well as between SNPs of STAT6 gene and the usefulness of topical nanoparticle cream (51).
Conclusions
The present study identified a functional SNP of TGFB1 to be associated with eczema susceptibility in southern Chinese children. We also found epistatic interactions among multiple immune regulatory genes for the eczema subphenotype total IgE levels, supporting the complex nature of eczema that involves dysregulated T cell immunity in children. This may be further studied in children of other ethnicities.
Acknowledgments
We thank Susan Wang, Hing Yee Sy, Gary Ching, Kathy Tsang, Kin Yee Wong, and Air Chan for helping to archive and process subjects’ blood samples and perform allergy testing.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR and STROBE reporting checklists. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-474/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-474/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-474/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-474/coif). T.F.L. serves as an unpaid editorial board member of Translational Pediatrics from March 2022 to February 2024. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee approved this study (Nos. 2008.123 and 2016.171). Subjects and/or their parents gave informed written consent to participate in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shin YH, Hwang J, Kwon R, et al. Global, regional, and national burden of allergic disorders and their risk factors in 204 countries and territories, from 1990 to 2019: A systematic analysis for the Global Burden of Disease Study 2019. Allergy 2023;78:2232-54. [Crossref] [PubMed]
- Global, regional, and national incidence of six major immune-mediated inflammatory diseases: findings from the global burden of disease study 2019. EClinicalMedicine 2023;64:102193. [Crossref] [PubMed]
- Damiani G, Calzavara-Pinton P, Stingeni L, et al. Italian guidelines for therapy of atopic dermatitis-Adapted from consensus-based European guidelines for treatment of atopic eczema (atopic dermatitis). Dermatol Ther 2019;32:e13121. [Crossref] [PubMed]
- Hammond M, Gamal A, Mukherjee PK, et al. Cutaneous dysbiosis may amplify barrier dysfunction in patients with atopic dermatitis. Front Microbiol 2022;13:944365. [Crossref] [PubMed]
- Damiani G, Finelli R, Kridin K, et al. Facial atopic dermatitis may be exacerbated by masks: insights from a multicenter, teledermatology, prospective study during COVID-19 pandemic. Ital J Dermatol Venerol 2022;157:505-9. [Crossref] [PubMed]
- Zhang J, Jiang P, Zhang Y, et al. Effects of wearing masks during COVID-19 pandemic on the composition and diversity of skin bacteria and fungi in medical workers. Front Microbiol 2023;14:1274050. [Crossref] [PubMed]
- Glatz M, Bosshard PP, Hoetzenecker W, et al. The Role of Malassezia spp. in Atopic Dermatitis. J Clin Med 2015;4:1217-28. [Crossref] [PubMed]
- Shi B, Bangayan NJ, Curd E, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol 2016;138:1233-6. [Crossref] [PubMed]
- Corry DB, Kheradmand F. Induction and regulation of the IgE response. Nature 1999;402:B18-23. [Crossref] [PubMed]
- Hamilton JD, Suárez-Fariñas M, Dhingra N, et al. Dupilumab improves the molecular signature in skin of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol 2014;134:1293-300. [Crossref] [PubMed]
- O'Regan GM, Sandilands A, McLean WHI, et al. Filaggrin in atopic dermatitis. J Allergy Clin Immunol 2008;122:689-93. [Crossref] [PubMed]
- He JQ, Chan-Yeung M, Becker AB, et al. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun 2003;4:385-9. [Crossref] [PubMed]
- Ching GK, Hon KL, Ng PC, et al. Filaggrin null mutations in childhood atopic dermatitis among the Chinese. Int J Immunogenet 2009;36:251-4. [Crossref] [PubMed]
- Noda S, Suárez-Fariñas M, Ungar B, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol 2015;136:1254-64. [Crossref] [PubMed]
- Moses HL. TGF-beta regulation of epithelial cell proliferation. Mol Reprod Dev 1992;32:179-84. [Crossref] [PubMed]
- Geherin SA, Gómez D, Glabman RA, et al. IL-10+ Innate-like B Cells Are Part of the Skin Immune System and Require α4β1 Integrin To Migrate between the Peritoneum and Inflamed Skin. J Immunol 2016;196:2514-25. [Crossref] [PubMed]
- Poulsen LK, Hummelshoj L. Triggers of IgE class switching and allergy development. Ann Med 2007;39:440-56. [Crossref] [PubMed]
- El Kasmi KC, Holst J, Coffre M, et al. General nature of the STAT3-activated anti-inflammatory response. J Immunol 2006;177:7880-8. [Crossref] [PubMed]
- Wang SS, Hon KL, Kong AP, et al. Eczema phenotypes are associated with multiple vitamin D pathway genes in Chinese children. Allergy 2014;69:118-24. [Crossref] [PubMed]
- Leung TF, Ching KW, Sy HY, et al. CHIA confers susceptibility to childhood eczema. Br J Dermatol 2010;163:1360-2. [Crossref] [PubMed]
- Ozaki R, Qiao Q, Wong GW, et al. Overweight, family history of diabetes and attending schools of lower academic grading are independent predictors for metabolic syndrome in Hong Kong Chinese adolescents. Arch Dis Child 2007;92:224-8. [Crossref] [PubMed]
- Wang SS, Hon KL, Sy HY, et al. Interactions between genetic variants of FLG and chromosome 11q13 locus determine susceptibility for eczema phenotypes. J Invest Dermatol 2012;132:1930-2. [Crossref] [PubMed]
- Chan NP, Choi KC, Nelson EA, et al. Associations of pubertal stage and body mass index with cardiometabolic risk in Hong Kong Chinese children: A cross-sectional study. BMC Pediatr 2015;15:136. [Crossref] [PubMed]
- Leung TF, Tang MF, Leung ASY, et al. Cadherin-related family member 3 gene impacts childhood asthma in Chinese children. Pediatr Allergy Immunol 2020;31:133-42. [Crossref] [PubMed]
- Wang SS, Hon KL, Kong AP, et al. Vitamin D deficiency is associated with diagnosis and severity of childhood atopic dermatitis. Pediatr Allergy Immunol 2014;25:30-5. [Crossref] [PubMed]
- Kong AP, Ko GT, Ozaki R, et al. Metabolic syndrome by the new IDF criteria in Hong Kong Chinese adolescents and its prediction by using body mass index. Acta Paediatr 2008;97:1738-42. [Crossref] [PubMed]
- Wildi K, Livingstone S, Palmieri C, et al. The discovery of biological subphenotypes in ARDS: a novel approach to targeted medicine? J Intensive Care 2021;9:14. [Crossref] [PubMed]
- Celakovská J, Bukac J, Ettler K, et al. Evaluation of Peripheral Blood Eosinophilia in Adolescent and Adult Patients Suffering from Atopic Dermatitis and the Relation to the Occurrence of Allergy to Aeroallergens. Indian J Dermatol 2019;64:34-40. [Crossref] [PubMed]
- Wärnberg Gerdin S, Lie A, Asarnoj A, et al. Impaired skin barrier and allergic sensitization in early infancy. Allergy 2022;77:1464-76. [Crossref] [PubMed]
- Loo EX, Sim JZ, Goh A, et al. Predictors of allergen sensitization in Singapore children from birth to 3 years. Allergy Asthma Clin Immunol 2016;12:56. [Crossref] [PubMed]
- Bottema RW, Kerkhof M, Reijmerink NE, et al. Gene-gene interaction in regulatory T-cell function in atopy and asthma development in childhood. J Allergy Clin Immunol 2010;126:338-46, 346.e1-10.
- Huang S, Wang H, Zheng H, et al. Association between IL-6 polymorphisms and Atopic Dermatitis in Chinese Han children. Front Pediatr 2023;11:1156659. [Crossref] [PubMed]
- Sumiyoshi K, Nakao A, Ushio H, et al. Transforming growth factor-beta1 suppresses atopic dermatitis-like skin lesions in NC/Nga mice. Clin Exp Allergy 2002;32:309-14. [Crossref] [PubMed]
- Arkwright PD, Chase JM, Babbage S, et al. Atopic dermatitis is associated with a low-producer transforming growth factor beta(1) cytokine genotype. J Allergy Clin Immunol 2001;108:281-4. [Crossref] [PubMed]
- Shah R, Hurley CK, Posch PE. A molecular mechanism for the differential regulation of TGF-beta1 expression due to the common SNP -509C-T (c. -1347C > T). Hum Genet 2006;120:461-9. [Crossref] [PubMed]
- Nagpal K, Sharma S. TGFbeta1 haplotypes and asthma in Indian populations. J Allergy Clin Immunol 2005;115:527-33. [Crossref] [PubMed]
- Panek M, Pietras T, Fabijan A, et al. Identification and association of the single nucleotide polymorphisms, C-509T, C+466T and T+869C, of the TGF-β1 gene in patients with asthma and their influence on the mRNA expression level of TGF-β1. Int J Mol Med 2014;34:975-86. [Crossref] [PubMed]
- Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, et al. TGFβ receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med 2013;5:195ra94. [Crossref] [PubMed]
- Hunninghake GM, Soto-Quirós ME, Lasky-Su J, et al. Dust mite exposure modifies the effect of functional IL10 polymorphisms on allergy and asthma exacerbations. J Allergy Clin Immunol 2008;122:93-8, 98.e1-5.
- Guo J, He YH, Chen F, et al. The A to G polymorphism at -1082 of the interleukin-10 gene is rare in the Han Chinese population. Mol Med Rep 2012;6:894-6. [Crossref] [PubMed]
- Lopez-Lasanta M, Julià A, Maymó J, et al. Variation at interleukin-6 receptor gene is associated to joint damage in rheumatoid arthritis. Arthritis Res Ther 2015;17:242. [Crossref] [PubMed]
- Gigante B, Strawbridge RJ, Velasquez IM, et al. Analysis of the role of interleukin 6 receptor haplotypes in the regulation of circulating levels of inflammatory biomarkers and risk of coronary heart disease. PLoS One 2015;10:e0119980. [Crossref] [PubMed]
- Esparza-Gordillo J, Schaarschmidt H, Liang L, et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J Allergy Clin Immunol 2013;132:371-7. [Crossref] [PubMed]
- Wang XP, Schunck M, Kallen KJ, et al. The interleukin-6 cytokine system regulates epidermal permeability barrier homeostasis. J Invest Dermatol 2004;123:124-31. [Crossref] [PubMed]
- Schaarschmidt H, Ellinghaus D, Rodríguez E, et al. A genome-wide association study reveals 2 new susceptibility loci for atopic dermatitis. J Allergy Clin Immunol 2015;136:802-6. [Crossref] [PubMed]
- Tang MF, Sy HY, Kong AP, et al. Genetic effects of multiple asthma loci identified by genomewide association studies on asthma and spirometric indices. Pediatr Allergy Immunol 2016;27:185-94. [Crossref] [PubMed]
- Leung TF, Wong CK, Chan IH, et al. Plasma concentration of thymus and activation-regulated chemokine is elevated in childhood asthma. J Allergy Clin Immunol 2002;110:404-9. [Crossref] [PubMed]
- Leung TF, Wong YS, Chan IH, et al. Indoor determinants of endotoxin and dust mite exposures in Hong Kong homes with asthmatic children. Int Arch Allergy Immunol 2010;152:279-87. [Crossref] [PubMed]
- Leung TF, Wong YS, Chan IH, et al. Domestic exposure to aeroallergens in Hong Kong families with asthmatic children. Pediatr Pulmonol 2011;46:632-9. [Crossref] [PubMed]
- Alroobaea R, Rubaiee S, Hanbazazah AS, et al. IL-4/13 Blockade and sleep-related adverse drug reactions in over 37,000 Dupilumab reports from the World Health Organization Individual Case Safety reporting pharmacovigilance database (VigiBase™): a big data and machine learning analysis. Eur Rev Med Pharmacol Sci 2022;26:4074-81. [Crossref] [PubMed]
- Damiani G, Eggenhöffner R, Pigatto PDM, et al. Nanotechnology meets atopic dermatitis: Current solutions, challenges and future prospects. Insights and implications from a systematic review of the literature. Bioact Mater 2019;4:380-6. [Crossref] [PubMed]


