
A rare inherited homozygous missense variant in PLA2G6 influences susceptibility to infantile neuroaxonal dystrophy: a case report
Highlight box
Key findings
• We report an infantile neuroaxonal dystrophy (INAD) child with a rare PLA2G6 c.1778C>T homozygous missense variant and associated clinical symptoms.
What is known and what is new?
• INAD is an ultra-rare early-onset neurological disorder caused by PLA2G6 variants. However, genotype-phenotype association is still lacking for a majority of PLA2G6 variants.
• We enrolled a 16-month-old boy with classical INAD neuropathological symptoms and his family members. By chromosome microarray analysis, whole-exome sequencing, and Sanger sequencing, we identified a PLA2G6 c.1778C>T homozygous missense variant in the child. By family-based cosegregation analysis, we confirmed that the PLA2G6 c.1778C>T homozygous variant contributes to the pathogenesis of INAD.
What is the implication, and what should change now?
• Our study emphasizes that genetic testing is essential for the early and accurate identification of INAD and reveals that PLA2G6 c.1778C>T homozygous variant contributes to the pathogenesis of INAD.
Introduction
Infantile neuroaxonal dystrophy (INAD; OMIM # 256600), originally named Seitelberger’s disease, is an ultra-rare early-onset neurological disorder with an autosomal recessive manner of inheritance (1-3). INAD’s prevalence is unknown, but its clinical manifestations have slowly been recognized from the cases described so far. Infants with typical INAD usually experience a short asymptomatic period after birth (4). Most of them, from the age of 6 months or even earlier, consistently develop multiple neuropathological symptoms, including but not limited to progressive psychomotor deterioration, mental retardation, axial hypotonia, cerebellar ataxia, hearing loss (HL), and optic nerve abnormalities, that jointly lead to a persistent vegetative state and premature death, generally early in the first decade of life (4,5). There is growing evidence that INAD is primarily caused by various loss-of-function variants in the phospholipase A2 group VI (PLA2G6) gene on chromosome 22q, while genetic and phenotypic heterogeneity exists (6-10). Nowadays, the definitive diagnosis of INAD heavily relies on genetic confirmation of PLA2G6 status in combination with the INAD clinical phenotypes. To date, in the Human Gene Mutation Database (HGMD), more than 100 PLA2G6 variants, the vast majority of which are missense variants, have been associated with INAD (7). However, the association between an individual’s genetic makeup (genotype) and the clinical characteristics (phenotype), i.e., genotype-phenotype association, is still lacking for many INAD patients attributed to PLA2G6 variants.
Here, we report a 16-month-old boy with typical clinical symptoms of INAD. Chromosomal microarray analysis (CMA) detected several copy number-neutral regions of runs of homozygosity (ROH), which involve dozens of recessively inherited diseases related genes, including the PLA2G6 gene. By whole-exome sequencing (WES), we identified a rare homozygous missense variant in the exon 13 of PLA2G6, i.e., NM_003560.2: c.1778C>T, p.Pro593Leu (rs1451486649), which was further checked in the proband’s family by Sanger sequencing analysis. As a result, we found that asymptomatic members in the enrolled family were all heterozygous carriers of the PLA2G6 c.1778C>T variant. Our study emphasizes that genetic testing is essential for the early and accurate identification of INAD and reveals that PLA2G6 c.1778C>T homozygous variant contributes to the pathogenesis of INAD. We present this article in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-568/rc).
Case presentation
Clinical manifestation
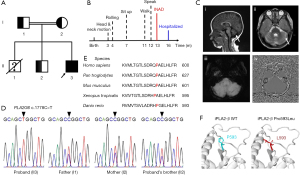
A 16-month-old boy (Figure 1A; II-3) was admitted to our hospital due to regression of acquired motor and speech abilities that had persisted for four months. The proband was born to a healthy consanguineous couple (Figure 1A; I-1, 30 years old; I-2, 30 years old) after 41 weeks of pregnancy and natural delivery. He had two brothers: the oldest brother (Figure 1A; II-1, 5 years old) died of respiratory failure caused by muscle dysfunction, and the other brother (Figure 1A; II-2, 5 years old) was healthy. At birth, the proband’s weight was 3.6 kg and height was 50 cm, and no obvious physical defects were noticed by his parents. He displayed head rotation and neck extension from 3 months of age; started rolling over at 4 months old; learned to sit up independently and walk with assistance at 7 and 11 months old, respectively. The first simple words “dada” and “mama” started at 12 months old. At that time, he was able to ask for something by pointing. Later on, the proband gradually lost his speech ability and showed lessened and stereotypic body movements. At the time of physical and neurological examination, he had a weight of 11 kg and a height of 82 cm. Notably, he failed to sit up by himself and developed astasia-abasia but could respond to “no” and simple instructions. The motor development timeline is summarized in Figure 1B. The electroencephalogram (EEG) showed normal activity. However, weakness of distal muscles and diminished patellar tendon reflex, but not pathologic reflexes, were observed. Brain magnetic resonance imaging (MRI) showed bilateral widening of the cerebellar sulci, indicating cerebellar atrophy, but brain iron accumulation was absent (Figure 1C). The auditory brainstem response (ABR) indicated moderately severe HL. His left ear had 60 decibels (dB) HL and the right ear had 70 dB HL. Moreover, the serum biochemical analysis showed a noticeable reduction in creatinine (19.7 µmol/L) and elevated levels of several enzymes, including aspartate aminotransferase (AST; 68 U/L), lactate dehydrogenase (LDH; 365.7 U/L), alkaline phosphatase (ALP; 135.6 U/L), and creatine kinase-myoglobin binding (CK-MB; 36.3 IU/L), and rest important indexes were within normal laboratory ranges (Table S1). Due to these clinical and laboratory findings, the proband was highly suspected with INAD, and this family was enrolled for further genetic analysis.
Genetic analysis
For the etiology screening of rare neurodevelopment disorders, especially for individuals born to consanguineous parents, chromosome microarray analysis (CMA) is the priority selection. Since CMA offers the capacity to detect ROH regions, in addition to copy number variants (CNVs) and single nucleotide polymorphisms (SNPs), for all chromosomes. Importantly, extensively existing ROH regions contain low-penetrance genes that are usually associated with recessively inherited diseases and increase susceptibility to them. Therefore, we performed CMA (Appendix 1), which revealed approximately 69 Mb ROH regions across the proband’s genome, but not CNVs. The ROH regions (>10 Mb) includes 1p21.3-p12 (19.09 Mb; 23 genes; 98,957,502–118,049,185 GRCh37), 4q31.21-q32.1 (14.41 Mb; 12 genes; 143,073,691–157,490,504 GRCh37), and 22q12.2-q13.31 (16.05 Mb; 21 genes; 31,245,745–47,297,593 GRCh37). Notably, PLA2G6 gene is located within the ROH region of 22q12.2-q13.31 (Table S2).
To make an accurate genetic diagnosis, we further implemented WES followed by Sanger sequencing in the investigated family (Appendix 1). After filtering out the common and non-coding variants, WES analysis found that the only homozygous variant that may reasonably explain INAD-like symptoms of the proband was NM_003560.2: c.1778C>T, p.Pro593Leu (rs1451486649) homozygous missense variant in PLA2G6 gene, while in several INAD-irrelevant genes, variants were all present in the heterozygous state (Table 1). Sanger sequencing confirmed the PLA2G6 homozygous variant in the proband and further revealed that the proband’s parents and alive sibling were all heterozygous for PLA2G6 c.1778C>T (Figure 1D). The identified rare PLA2G6 c.1778C>T was previously known as a likely pathologic variant with ClinVar star 2+ according to the American College of Medical Genetics and Genomics (ACMG) guidelines. PLA2G6 c.1778C>T has a total allele frequency of 3×10−6 in GnomAD, but it was not included in ExAC (Exome Aggregation Consortium), 1000 Genomes Project, HapMap, and our homemade exome database.
Table 1
| Gene | Reference sequence | Variant | Protein consequence | Alleles status | VEP annotation | Interpretation in ClinVar | Inherited pattern | Related disease (s) |
|---|---|---|---|---|---|---|---|---|
| PLA2G6 | NM_003560.2 | c.1778C>T | p.Pro593Leu | Homozygous | Missense | Likely pathogenic, uncertain significance | AR | Infantile neuroaxonal dystrophy 1 |
| Neurodegeneration with brain iron accumulation 2B | ||||||||
| Parkinson disease 14 | ||||||||
| TRIP12 | NM_004238.1 | c.592T>G | p.Ser198Ala | Heterozygous | Missense | Not reported | AD | Clark-Baraitser syndrome |
| TBR1 | NM_006593.2 | c.1351_1356dup | p.Ala451_Gly452dup | Heterozygous | Frameshift | Not reported | AD | Intellectual developmental disorder with autism and speech delay |
| ASXL3 | NM_030632.1 | c.2065A>C | p.Ile689Leu | Heterozygous | Missense | Benign | AD | Bainbridge-Ropers syndrome |
| VPS53 | NM_001128159.3 | c.1517G>A | p.Arg506Gln | Heterozygous | Missense | Uncertain significance | AR | Pontocerebellar hypoplasia, type 2E |
| HSD17B4 | NM_000414.3 | c.508G>A | p.Gly170Ser | Heterozygous | Missense | Not reported | AR | Perrault syndrome 1 |
| D-bifunctional protein deficiency | ||||||||
| PDE10A | NM_001130690.2 | c.62G>C | p.Ser21Thr | Heterozygous | Missense | Not reported | AR | Dyskinesia, limb and orofacial, infantile onset |
| AD | Striatal degeneration | |||||||
| ATP13A2 | NM_022089.2 | c.1447T>G | p.Tyr483Asp | Heterozygous | Missense | Not reported | AR | Spastic paraplegia 78 |
| Kufor-Rakeb syndrome |
WES, whole-exome sequencing; VEP, variant effect predictor; AR, autosomal recessive; AD, autosomal dominant.
Molecular analysis
Conservation analysis of PLA2G6-encoded protein, iPLA2-β, indicated that Pro593 is a highly conserved site from Danio rerio to Homo sapiens (Figure 1E). PLA2G6 c.1778C>T homozygous variant results in the replacement of the hydrophobic and cyclic amino acid proline (Pro593) located in the iPLA2-β catalytic domain (CAT) by another hydrophobic, but aliphatic, amino acid leucine (Pro593Leu) (Figure 1F). This missense variant is predicted to be pathogenic by functional analysis with SIFT (RRID:SCR_012813) (12), PolyPhen2 (RRID:SCR_013189) (13), and ClinPred (14). Thus, the proband was diagnosed with INAD, which was potentially caused by a rare inherited PLA2G6 c.1778C>T homozygous pathologic variant.
Ethical considerations
All procedures performed in this study were in accordance with the ethical standards of the Ethics Committee of the Huzhou Maternity & Child Health Care Hospital and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s legal guardians for the publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
The PLA2G6 gene, which encodes a Ca2+-independent phospholipase A2 (iPLA2-β), is located on chromosome 22q13.1 and contains 19 exons, including two alternative exons (15). PLA2G6 is a well-known pathogenic gene for a series of autosomal recessively inherited neurodegenerative diseases, mainly including INAD, atypical neuroaxonal dystrophy (AND), and early-onset parkinsonism (AREP) (7). Collectively, the term PLA2G6-associated neurodegeneration (PLAN) has been derived to describe all diseases resulting from PLA2G6 gene variants (7). Among PLAN, INAD is recognized by the earliest onset of psychomotor deterioration and the shortest life span (7). This phenomenon is likely attributed to the fact that INAD-related PLA2G6 variants result in more severe impairment of iPLA2-β enzymatic activity compared to PLA2G6 variants linked to other PLAN disorders (16). It is also noteworthy that the same PLA2G6 variant can lead to different clinical phenotypes among individuals (8,9,17,18). For example, compared to our INAD proband, patient (43-year-old man) with the PLA2G6 c.991G>T homozygous variant, which causes a 70% reduction in enzyme activity of iPLA2-β, developed AREP phenotypes with relatively later onset ages and slower disease progression (17). However, his younger sister (34 years old) with the same homozygous variant displayed no symptoms of PLAN (17).
In this study, our INAD proband carried PLA2G6 c.1778C>T homozygous variant, which causes a conserved proline substitution by leucine in the iPLA2-β CAT. The mammalian iPLA2-β harbors two CATs, which are widely recognized for their dual roles in regulating iPLA2-β enzymatic activity (19). On one hand, CATs harbor active sites directly engaged in the phospholipid hydrolysis reaction (19). On the other hand, they also contribute to the iPLA2-β dimerization process (19). Given that leucine lacks the cyclic structure found in proline, the Pro593Leu substitution has the potential to introduce conformational change(s) in CATs and impact iPLA2-β dimerization. In addition, leucine is aliphatic and typically more soluble in water than proline, which may influence the hydrophobicity of CATs and impact molecular interactions with active sites. Maybe due to aforementioned factors, predictive tools like SIFT, PolyPhen2, and ClinPred have all indicated that PLA2G6 c.1778C>T variant to be deleterious. ClinVar database has included PLA2G6 c.1778C>T variant and defines it as a likely pathologic variant. Actually, in 2016, Al-Maawali et al. first reported the co-occurrence of PLA2G6 c.1778C>T variant and PLA2G6 c.1974C>A variant in an INAD patient (20). Nevertheless, limited clinical information of this patient was described (20). From this solely available report, one can hardly make an accurate assessment of the contribution of PLA2G6 c.1778C>T to the pathogenesis of INAD. In this study, we detected the PLA2G6 c.1778C>T homozygous variant in a 16-month-old boy and described his INAD clinical characterization in detail. His inbred parents and alive sibling were all asymptomatic and, importantly, heterozygous for PLA2G6 c.1778C>T. These results clearly suggest that the proband’s PLA2G6 c.1778C>T homozygous variant is recessively inherited from both parents and leads to INAD.
Before the discovery of the linkage between PLA2G6 variants and INAD pathogenesis in 2006 (2,3), the diagnosis of INAD mainly relied on a spectrum of unspecific clinical phenotypes. However, the most initial noticeable INAD symptoms, i.e., psychomotor regression and muscle weakness may also lead to a diagnosis of other neurodevelopment disorders, such as hereditary spastic paraplegia (HSP) (21). In most patients, brain MRI detects cerebellar atrophy early in INAD. As INAD progresses, on T2-weighted MRI, gradient echo (GRE), or susceptibility-weighted imaging (SWI), abnormal low signals commonly appear primarily in the globus pallidus, indicating iron accumulation. However, cerebellar atrophy and iron accumulation also can be found in several other diseases (6). Besides, iron accumulation appears only at the late stage of INAD (4,5). In the patient with PLA2G6 c.1778C>T/c.1974C>A compound heterozygous variants, brain iron accumulation was absent at age of 2 years (20). In consistent with this, we also failed to detect iron accumulation in our patient at the age of 16 months. Thus, MRI provides limited benefit for early INAD diagnosis. Notably, in PLA2G6-mutant patients, the observation of elevated AST and LDH levels (22,23), which were confirmed in our proband, raises the possibility that these two enzymes represent potential biomarkers of INAD. Our findings also indicated reduced creatinine levels and elevated ALP/CK-MB levels in the proband, indicating a widespread enzyme metabolism disorder and, possibly, kidney, heart, and multiple tissue damage. Furthermore, axon spheroids and vacuoles revealed by peripheral nerve biopsies have long been regarded as a gold standard for INAD diagnosis, with an estimated 87% of PLA2G6 mutation-positive INAD having axon spheroids (5). However, parents of INAD children have a less than enthusiastic response to cooperating with peripheral nerve biopsies, once they are informed of its invasive procedures, at least in our case.
To date, treatments of INAD and other PLAN are still palliative. Enzyme replacement therapy and gene therapy, as innovative approaches to rectify iPLA2-β enzyme activity, are still in the early stages of development (1). In our case, the proband’s parents decided to refrain from any treatment because of the similar disease experience of their first child. We provided the couple with counseling on in vitro fertilization (IVF), preimplantation genetic diagnosis (PGD), and prenatal diagnosis if they wanted to have another child.
It is crucial to highlight that the proband harbored four heterozygous variants in pathogenic genes associated with autosomal dominant (AD) diseases (Table 1). While ASXL3 c.2065A>C (NM_030632.1; rs563644271) is classified as a benign variant in ClinVar, the interpretations of the remaining variants have not been documented. Notably, heterozygous variants in the TRIP12, TBR1, and PDE10A are known to cause Clark-Baraitser syndrome (CLABARS; OMIM # 617752), intellectual development with autism and speech delay (IDDAS; OMIM # 606053), and autosomal-dominant striatal degeneration (ADSD; OMIM # 616922), respectively (24-26). All the three rare diseases show neurological symptoms or signs. In general, CLABARS is characterized by intellectual disability, developmental delay, macrocephaly, and distinct facial deformities (24). Autism spectrum disorder and delayed speech and intellectual functions are hallmark signs of IDDAS (25). Clinical symptoms of ADSD include slowly progressive dysarthria, gait disturbance, muscle rigidity, and dysdiadochokinesia (26). At clinical level, progressive neurological regression and the absence of physical deformities stand out as the distinguishing features of INAD when compared to CLABARS and IDDAS. Muscle weakness and hypotonia are distinctive features that set apart INAD from ADSD in affected patients. Meanwhile, onset and age of presentation are also helpful for differential diagnosis. Even though the enrolled family did not exhibit the classic symptoms of CLABARS, IDDAS, and ADSD, we cannot dismiss the possibility that TRIP12, TBR1, and PDE10A variants are de novo, which may predispose the proband to those AD diseases in the future.
We acknowledge several limitations of the present study. First, we did not perform peripheral nerve biopsies to examine axon spheroids. Second, we cannot exclude the possibility that multiple heterozygous variants (Table 1) may also contribute to the proband’s symptoms. Furthermore, the tracking time of the proband was short, making clinical characterization of INAD progression difficult. From the latest telephone follow-up, we were informed that the proband only made tiny movements in bed at the age of 2.5 years. Ultimately, more research is required to determine the mechanism by which the PLA2G6 c.1778C>T homozygous variant alters the activity of iPLA2-β and leads to INAD.
Conclusions
In summary, we reported detailed clinical characteristics of an INAD patient bearing PLA2G6 c.1778C>T homozygous variant. Our study expands the genetic and clinical spectrum of PLA2G6 pathogenic variants and provides important information about the pathogenesis of PLA2G6-related INAD.
Acknowledgments
We are grateful to the patient and his family, as well as the help of all the physicians in the course of the medical care.
Funding: This work was supported by grants from
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-568/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-568/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-568/coif). Y.L. receives funding from Huzhou Science and Technology Project of Zhejiang Province (Funding No. 2022GYB54). T.W. is a current employee of Dian Diagnostics Group Co., Ltd., Hangzhou, Zhejiang Province. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the Ethics Committee of the Huzhou Maternity & Child Health Care Hospital and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s legal guardians for the publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Babin PL, Rao SNR, Chacko A, et al. Infantile Neuroaxonal Dystrophy: Diagnosis and Possible Treatments. Front Genet 2018;9:597. [Crossref] [PubMed]
- Morgan NV, Westaway SK, Morton JE, et al. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat Genet 2006;38:752-4. Erratum in: Nat Genet 2006;38:957. [Crossref] [PubMed]
- Khateeb S, Flusser H, Ofir R, et al. PLA2G6 mutation underlies infantile neuroaxonal dystrophy. Am J Hum Genet 2006;79:942-8. [Crossref] [PubMed]
- Altuame FD, Foskett G, Atwal PS, et al. The natural history of infantile neuroaxonal dystrophy. Orphanet J Rare Dis 2020;15:109. [Crossref] [PubMed]
- Deng X, Yuan L, Jankovic J, et al. The role of the PLA2G6 gene in neurodegenerative diseases. Ageing Res Rev 2023;89:101957. [Crossref] [PubMed]
- Gregory A, Kurian MA, Maher ER, et al. PLA2G6-Associated Neurodegeneration. In: Adam MP, Feldman J, Mirzaa GM, et al. editors. GeneReviews®. Seattle, WA, USA: University of Washington, Seattle, 2008.
- Guo YP, Tang BS, Guo JF. PLA2G6-Associated Neurodegeneration (PLAN): Review of Clinical Phenotypes and Genotypes. Front Neurol 2018;9:1100. [Crossref] [PubMed]
- Rostampour D, Zolfaghari MR, Gholami M. Novel insertion mutation in the PLA2G6 gene in an Iranian family with infantile neuroaxonal dystrophy. J Clin Lab Anal 2022;36:e24253. [Crossref] [PubMed]
- Ansari B, Nasiri J, Namazi H, et al. Infantile Neuroaxonal Dystrophy in Two Cases: Siblings with Different Presentations. Iran J Child Neurol 2022;16:193-8. [PubMed]
- Zou Y, Luo H, Yuan H, et al. Identification of a Novel Nonsense Mutation in PLA2G6 and Prenatal Diagnosis in a Chinese Family With Infantile Neuroaxonal Dystrophy. Front Neurol 2022;13:904027. [Crossref] [PubMed]
- Khanna T, Hanna G, Sternberg MJE, et al. Missense3D-DB web catalogue: an atom-based analysis and repository of 4M human protein-coding genetic variants. Hum Genet 2021;140:805-12. [Crossref] [PubMed]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812-4. [Crossref] [PubMed]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7:Unit7.20.
- Alirezaie N, Kernohan KD, Hartley T, et al. ClinPred: Prediction Tool to Identify Disease-Relevant Nonsynonymous Single-Nucleotide Variants. Am J Hum Genet 2018;103:474-83. [Crossref] [PubMed]
- Larsson Forsell PK, Kennedy BP, Claesson HE. The human calcium-independent phospholipase A2 gene multiple enzymes with distinct properties from a single gene. Eur J Biochem 1999;262:575-85. [Crossref] [PubMed]
- Engel LA, Jing Z, O'Brien DE, et al. Catalytic function of PLA2G6 is impaired by mutations associated with infantile neuroaxonal dystrophy but not dystonia-parkinsonism. PLoS One 2010;5:e12897. [Crossref] [PubMed]
- Shi CH, Tang BS, Wang L, et al. PLA2G6 gene mutation in autosomal recessive early-onset parkinsonism in a Chinese cohort. Neurology 2011;77:75-81. [Crossref] [PubMed]
- Chu YT, Lin HY, Chen PL, et al. Genotype-phenotype correlations of adult-onset PLA2G6-associated Neurodegeneration: case series and literature review. BMC Neurol 2020;20:101. [Crossref] [PubMed]
- Malley KR, Koroleva O, Miller I, et al. The structure of iPLA(2)β reveals dimeric active sites and suggests mechanisms of regulation and localization. Nat Commun 2018;9:765. [Crossref] [PubMed]
- Al-Maawali A, Yoon G, Feigenbaum AS, et al. Validation of the finding of hypertrophy of the clava in infantile neuroaxonal dystrophy/PLA2G6 by biometric analysis. Neuroradiology 2016;58:1035-42. [Crossref] [PubMed]
- Murala S, Nagarajan E, Bollu PC. Hereditary spastic paraplegia. Neurol Sci 2021;42:883-94. [Crossref] [PubMed]
- Dastsooz H, Nemati H, Fard MAF, et al. Novel mutations in PANK2 and PLA2G6 genes in patients with neurodegenerative disorders: two case reports. BMC Med Genet 2017;18:87. [Crossref] [PubMed]
- Kraoua I, Romani M, Tonduti D, et al. Elevated aspartate aminotransferase and lactate dehydrogenase levels are a constant finding in PLA2G6-associated neurodegeneration. Eur J Neurol 2016;23:e24-5. [Crossref] [PubMed]
- Aerden M, Denommé-Pichon AS, Bonneau D, et al. The neurodevelopmental and facial phenotype in individuals with a TRIP12 variant. Eur J Hum Genet 2023;31:461-8. [Crossref] [PubMed]
- McDermott JH, Study DDD, Clayton-Smith J, et al. The TBR1-related autistic-spectrum-disorder phenotype and its clinical spectrum. Eur J Med Genet 2018;61:253-6. [Crossref] [PubMed]
- Mencacci NE, Kamsteeg EJ, Nakashima K, et al. De Novo Mutations in PDE10A Cause Childhood-Onset Chorea with Bilateral Striatal Lesions. Am J Hum Genet 2016;98:763-71. [Crossref] [PubMed]


