
CABP4 mutation in mice shows alteration in protein expression level and neuron discharge frequency
Highlight box
Key findings
• The Western blot results showed a significant difference in the brain stem, compared to the cortex, basal ganglia, and hippocampus area, in the mutant mice.
What is known, and what is new?
• We examined Chinese autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) pedigree and found that the calcium-binding protein 4 (CABP4) mutation might be linked to ADNFLE.
• We provided further evidence that CABP4 could be an epilepsy-related gene.
What is the implication, and what should change now?
• More experiments should be conducted with animal models, such as behavior experiments and epilepsy phenotype evaluations, to evaluate the phenotype of this mutation, and provide further evidence that CABP4 gene is a new pathogenic gene of ADNFLE.
Introduction
Autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE), also now known as autosomal dominant sleep-related hypermotor epilepsy, is the first type of idiopathic focal epilepsy for which specific mutations have been described. ADNFLE is characterized by hypermotor seizures arising predominantly during non-rapid eye movement sleep, and usually starts in childhood (1).
The ADNFLE disorder was first reported in 1994 (2), and the first ADNFLE-related gene, cholinergic receptor nicotinic alpha 4 subunit (CHRNA4), was first identified in an Australian pedigree in 1995 (3). With advancements in the development and application of next-generation sequencing technologies, more and more genes that might be related to ADNFLE have been reported, such as the genes encoding cholinergic receptor nicotinic beta 2 subunits (CHRNB2) (4) and alpha 2 subunits (CHRNA2) (5), the gene encoding the component of the GATOR1 complex (DEPDC5) (6), the gene encoding potassium sodium-activated channel (KCNT1) (7), the gene encoding corticotropin-releasing hormone (CRH) (8), and the gene encoding the calcium-binding protein 4 (CABP4) (9).
To date, the CABP4 gene has mainly been reported in autosomal recessive incomplete congenital stationary night blindness (10). CABP4 is specifically expressed at the synaptic terminals of photoreceptors in rods and cones, and it colocalizes and interacts with the alpha-1F subunit of L-type calcium voltage-dependent channels (Cav1.4), thereby regulating the calcium influx, which is important for maintaining normal synaptic function and morphology (11). CABP4-associated epilepsy was first reported in a Chinese family, in which 11 of 27 family members were diagnosed with ADNFLE without night blindness or visual disorders (9). Whole-exome sequencing was performed on 15 of the family members, of whom seven were affected and eight were unaffected. The CABP4 heterozygous (HE) missense mutation occurs at genetic sequence position 464. In this mutation, the guanine nucleotide is replaced by adenine, which leads to the amino acid glycine being replaced by aspartic acid at protein position 155 (c.464G>A, p.G155D). This mutation was detected in all the seven affected individuals and one unaffected family member. In vitro, human hippocampal neurons carrying the mutation showed increased CABP4 messenger RNA (mRNA) expression and decreased CABP4 protein production (12). All previous experiments on epileptogenicity of the CABP4 mutation were only based on in vitro cellular expression systems and thus cannot fully elucidate the effect of the mutation on neural networks and cerebral structures. To extend understandings of the mechanism underlying this type of epilepsy, we constructed a knock-in genetic mouse strain carrying the CABP4 c.464G>A mutation to elucidate the molecular, cellular, and neurophysiological basis of CABP4-associated epilepsy. Our findings might shed more light on the diagnosis and treatment of epilepsy. We present this article in accordance with the ARRIVE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-484/rc).
Methods
Generation of the CABP4G155D/+ mutant knock-in mouse model
A CABP4G155D/+ knock-in C57BL/6J mouse model was established using the CRISPR/Cas9 system at the Shanghai Model Organisms Center, Inc. The single-guide RNA (sgRNA) was designed to target the sequence 5'-TACATCGGCTACCGCGAGCT-3'. Designed Cas9 mRNA, sgRNA, and donor vectors carrying the mutant DNA fragment were co-injected into the C57BL/6J mouse zygotes. The system cleaved a double-strand break on the target sequence and repaired it by homologous recombination with donor vectors. Blood samples for sequencing were collected from the tails of potential founders to confirm the genotype. Genotypes were identified by amplified fragment and sequencing, using the primers 5'-CTGGAGGAGCGGTGGCAAAT-3' and 5'-TCCCATCCCCTGCTTAGAAAC-3', and the sequencing primer 5'-CTGGAGGAGCGGTGGCAAAT-3'.
To breed the mutant mice with stable hereditary ability, we crossed founders with wild-type (WT) C57BL/6J mice and generated the F1 generation of CABP4G155D/+ HE mice. Tail-tip tissues were collected at 3 weeks old for genotype analysis. According to the results, the animals were divided into the following two groups: the WT group; and the HE group.
A protocol, which was not registered, was established before the study commenced. All animals of the same-sex were housed in plastic cages (five animals per cage), with a constant temperature of 22±2 °C and a humidity of 60%±10%, maintained under a 12-h dark/light cycle, with free access to water and food. After birth, physical growth data, including body length, weight, and tail length, were collected weekly for 5 weeks.
All the animal-related experimental procedures were performed under a project license (No. 2018003A) granted by the Ethics Committee for Medical Research, Guangdong Provincial People’s Hospital, in compliance with the Chinese National Regulations on the Administration of Laboratory Animals and the Guangdong Provincial People’s Hospital Guideline of Ethical Use and Care of Experimental Animals.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Mature WT (n=3) and HE (n=3) mice were chosen randomly. The brains of both genotypes were quickly removed. Both cortex and thalamus tissues were collected and ground to isolate RNA with 1 mL of Trizol reagent (R0016; Beyotime Biotechnology, Shanghai, China) in accordance with the manufacturer’s protocol. RNA from each sample was reverse-transcribed to complementary DNA. qRT-PCR was performed under the following conditions: 95 °C (30 seconds; initial denaturing), 40 cycles of 95 °C (5 seconds; denaturing), 60 °C (30 seconds; annealing), and 60 °C (30 seconds; extension); and 72 °C (for 5 minutes to stop the procedure). The amplification curve and dissolution curve were used to verify the specificity of the amplified product. The comparative threshold cycle method (2−ΔΔCt) was used to calculate the fold-change expression of a particular gene, and the mouse glyceraldehyde-3-phosphate dehydrogenase gene was set as the reference standard. The data analysis procedure was performed by the ABI Prism 7300 SDS System (Applied Biosystem, Thermo Fisher Scientific, Waltham, MA, USA).
Western blot
Mature WT (n=3) and HE (n=3) mice were chosen randomly. Tissues were isolated and collected from the cortex, brain stem, basal ganglia, and hippocampus of mouse brains from the two groups. The samples were homogenized in radioimmunoprecipitation assay lysis buffer (P0013B; Beyotime Biotechnology) and protein was extracted using protease inhibitor phenylmethanesulfonylfluoride (PMSF) (04693132001; Roche Life Science, Rotkreuz, Switzerland), centrifuged at 12,000 rounds per minute at 4 °C for 15 minutes, and the upper clear liquid was then collected. The total amount of protein was determined using the bicinchoninic acid protein assay kit (P0012; Beyotime Biotechnology) in accordance with the manufacturer’s protocol. An equal amount of sample protein (80 µg) was loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. After electrophoresis, the sample was transformed onto a polyvinylidene difluoride membranes and blocked with 5% non-fat blocking grade milk (Huaxing Biotechnology, Beijing, China) for 1 hour at room temperature. Next, the membranes was incubated with the following primary antibodies overnight at 4 °C: anti-CABP4 (1:1,500, Santa Cruz Biotechnology, Dallas, USA), and anti-β-actin (1:5,000, Santa Cruz Biotechnology). Appropriate secondary antibodies (1:10,000, Thermo Fisher Scientific) were selected and incubated with the membrane for 1 hour at room temperature. After the incubation, the blot bands were visualized with an enhanced chemiluminescence reagent and quantified by densitometry using ImageJ software (version 1.51, National Institutes of Health, Bethesda, MD, USA). The results were normalized using β-actin as an internal control.
Patch-clamp analysis
WT (n=3) and HE (n=3) C57BL/6J male mice, both 8 weeks of age, were chosen for the brain slice patch-clamp analysis. After anesthetizing and decapitating the animal, the brain was harvested and, quickly and gently placed in oxygenated iced artificial cerebrospinal fluid (ACSF) (with 234 mM of sucrose, 2.5 mM of KCl, and 1.25 mH of NaH2PO4·2H2O). The cerebellum was removed carefully. The remaining brain tissue was fixed on agarose gel blocks with a fast-drying glue and placed carefully on the stage of a vibrate tissue slicer (VT 1000S; Leica, Wetzlar, Germany). Prefrontal lobe brain slices (300 µm in thickness) were cut and the slices were transferred into the recording ACSF (with 125 mM of NaCl, 2.5 mM of KCl, 1.25 mM of NaH2PO4·2H2O, 25 mM of NaHCO3, 10 mM of D-glucose, 2 mM of CaCl2·2H2O, and 1.5 mM of MgSO4) at a constant temperature of 32 °C for 30 minutes. One brain slice was placed in the recording chamber with ACSF at a constant temperature of 31±1 °C, and the flow was adjusted to 6 mL/min. The micropipette was pulled using borosilicate glass tubes (BF150-11-10; Sutter Instrument, Novato, CA, USA) and pipette pullers (P97; Sutter Instrument). The 40× immersion water objective lens [numerical aperture (NA) 0.8] microscope (BX51-WI; Olympus, Tokyo, Japan) was used to observe the target cells. The internal solution [with 140 mM of K-gluconate, 2 mM of MgCl2, 10 mM of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, 8 mM of KCl, 2 mM of Na2-ATP, and 0.2 mM of Na2-GTP, with the pH adjusted to 7.3 by KOH] was filled in the recording micropipette. Next, 1 µM of tetrodotoxin (TTX) and 10 µM of bicuculline were mixed in the extracellular fluid. The micropipette and recording electrode were moved vertically toward the pyramidal cell in the CA2 and CA3 layer of the mouse’s prefrontal cortex to generate compensatory potential. The microscope was attached to the target cell membrane and switched to negative pressure to form a gigaohm seal (>1 GΩ). The input resistance was monitored every 30 seconds, and the micro-excitatory post-synaptic currents (mEPSCs) were recorded for 10 minutes when the input resistance was stable. The experiment data were collected and analyzed only when the series resistance (10–30 MΩ), input resistance (100–300 MΩ), and post-synaptic current were all stable and did not exceed 20% of the mean value during the control period, and the initial mEPSC amplification was greater than 100 pA. The data were recorded by pClamp 10.5 software and analyzed by Clampfit 10.6 software (Molecular Devices, San José, CA, USA).
Statistical analysis
All the data are presented as the mean ± standard error of the mean or the median. All the statistical analyses were performed using the SPSS 26.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism 9.0 software. The normality of the data was tested using the Shapiro-Wilk test. An unpaired t-test or one-way analysis of variance (ANOVA) was used if the normality test was passed, if not, the Mann-Whitney test or Kruskal-Wallis test was used. A P value <0.05 was considered statistically significant.
Results
No obvious difference in the appearance, growth, and development was observed between the two genotypes
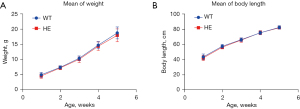
The CABP4G155D/+ mutant mouse model was successfully established, and the genotypes of all the animals were confirmed by Sanger sequencing (Figure 1). The animals were divided into two groups according to their genotype. Both the WT and HE mice were born at the expected Mendelian ratio. No obvious differences between the HE and WT mice were observed during the daily observations in terms appearance or abnormal behavior (e.g., spontaneous seizure), nor was any difference observed in terms of the weight and body length growth speeds. However, the average weight of the 1-week-old WT mice (4.993±0.785 g) was slightly heavier than that of the HE mice of the same age (4.596±0.757 g), but no statistically significant difference was observed in the 5-week-old mice (Figure 2A,2B).

A higher CABP4 protein level was observed in the mutant mouse brain stem tissue
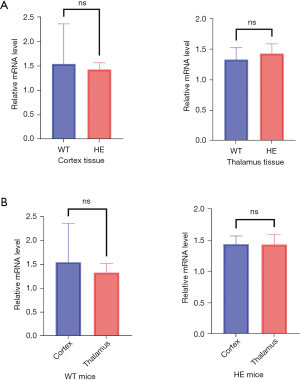
QRT-PCR was performed to evaluate the CABP4 mRNA expression levels in both the WT and HE mice. All the data were recorded and analyzed using the 2−ΔΔCt method. The Mann-Whitney U test was performed on GraphPad 9.0. No significant differences were found in the mRNA expression levels between the same brain region tissue in the two genome types or the two brain regions of the same genome type (Figure 3A,3B). This suggests that the mutation might not affect the mRNA expression level of CABP4 in vivo.
Western blot was performed to examine the CABP4 protein production level of the two groups. Tissue was collected from four different regions of mouse brains (i.e., the cortex, brain stem, basal ganglia, and hippocampus). β-actin was chosen as the internal reference gene (Figure 4A). The grey level of each band was analyzed by ImageJ software. Using SPSS 26.0 and GraphPad Prism 9.0, the unpaired t-test and a one-way ANOVA, followed by Tukey’s multiple comparisons tests, were used to analyze the standardized data, and a P value <0.05 was considered statistically significant. The analyzed results suggested that there was no significant difference in the CABP4 protein levels between the two different genotypes (Figure 4B). However, in the CABP4G155D/+ mice, the results indicated that the CABP4 protein level was higher in the brain stem than basal ganglia or hippocampus (P<0.05) (Figure 4C). Thus, the mutation could affect the CABP4 protein production levels in the different brain regions, which might be a factor leading to increased epilepsy susceptibility.

The prefrontal cortex layer II/III had a higher mEPSC discharge frequency
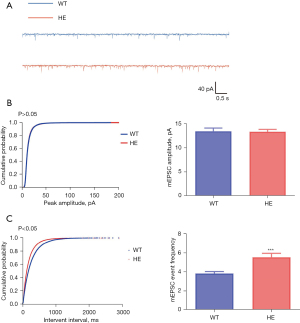
We selected pyramidal cells from layers II and III of the mouse’s prefrontal lobe cortex, as the ADNFLE patient’s electroencephalography (EEG) indicated that epilepsy usually started from the neocortex of the frontal lobe. After using TTX to block the action potential and cadmium to block the voltage-gated calcium channels, we recorded the peak amplitude and frequency of the mEPSCs in both genotype cells (Figure 5A). There was no significant difference between the amplitude of the two genotypes (AmplitudeWT =13.46±0.66, AmplitudeHE =13.36±0.49, P=0.90) (Figure 5B), but there was a significant difference in the average frequency (FrequencyWT =3.83±0.20, FrequencyHE =5.53±0.41, P<0.001) (Figure 5C). These results indicated that the mEPSC frequency in the pyramidal cells was higher in the HE mice than the WT mice, but no obvious difference in peak amplitude was observed.
Discussion
To further elucidate the relationship between CABP4 and epilepsy, a CABP4 (c.464G>A, p.G155D) HE mutation was introduced into C57BL/6J mice, and the mutant mice were then crossed with the WT mice to confirm the germline transmission of the mutation. The CABP4G115D/+ mice were born at the expected Mendelian ratio, which suggests that the mutation had little negative effect on perinatal survival. However, no spontaneous motor seizures were observed in the mutant mice as expected. Because the results in this study only represent simple cursory observations, which are short and scattered, and because of the effects of incomplete episodic episodes of the causative gene in ADNFLE, and the serendipitous nature of seizures, it is unclear whether the CABP4 HE mutation exhibits an epileptic phenotype. Thus, further behavioral evaluations, prolonged videotaped observations, and also assessments of changes in epilepsy susceptibility are needed in the future.
The CABP4 gene (OMIM: 608965) is a member of the calcium-binding protein family, and is highly similar to calmodulin both in structure and function (13). Previous studies have shown that CABP4 functions as a regular protein of voltage-dependent calcium channels Cav1.3 (14) and Cav1.4 (15), and plays an important role in synaptic function (16,17).
We also found that while the CABP4 (c.464G>A, p.G155D) HE mutation did not affect the overall expression of mRNA and protein, it did cause an imbalance in protein expression in the brain region. The brainstem connects the cerebrum, diencephalon, and spinal cord with complex neuronal networks, such as the ascending reticular activating system. It is thought to play an essential role in seizure initiation and propagation (18). A combined analysis of brainstem auditory evoked potentials and EEG of patients without epilepsy (19) and generalized convulsive seizure patients (20) revealed biphasic excitability changes in the brainstem, especially the lower brainstem. These excitability changes are conducted through the ascending activating system to the cortex, and produce a poly spike and wave complex. Brainstem lesions are also reported to be important in multiple sclerosis-related epilepsy (21).
The brainstem is closely related to both generalized epilepsy and focal epilepsy [i.e., temporal lobe epilepsy (TLE)]. In a case-control study of 26 TLE patients, magnetic resonance imaging showed ascending reticular activating system structural and functional connectivity reduction in the TLE patients, and the reduction was associated with the consciousness-impairment frequency and the presence of generalized seizures. Li also reported that brainstem raphe nucleus hypoechogenicity is related to spike frequency in epilepsy patients (22). Based on previous studies demonstrating the important role of CABP4 in synapse formation and development and that the brainstem has neural loops that maintain and generate epileptic states, we hypothesized that CABP4 (c.464G>A, p.G155D) HE mutations might be caused by abnormalities in the synaptic structure, which may lead to an increased susceptibility to epilepsy. Therefore, the brainstem appears to play an important role in CABP4-associated epilepsy.
To investigate the relative magnitudes of excitatory and inhibitory electrical signals, we examined the micro-excitatory post-synaptic membrane current signals in prefrontal cortex layer II or III pyramidal cells of WT and HE mice using the brain slice membrane clamp technique. The results revealed no significant difference in the current amplitude but a significant increase in the mEPSC frequency in the mutant mice. It is known that the magnitude of mEPSC, which represents post-synaptic events, depends on the number of post-synaptic receptors and the size of the pre-synaptic released vesicles (23). Conversely, the mEPSC frequency, represents the pre-synaptic event and is usually correlated with the probability of the change in the number of synapses and neurotransmitter releases (24). Based on the electrophysiological results of the present study, the CABP4 (c.464G>A, p.G155D) HE mutant mice appeared to spontaneously release more excitatory pre-synaptic vesicles than the WT mice. Thus, the CABP4 mutation may increase the release of excitatory neurotransmitters, which can disrupt the delicate balance of excitatory and inhibitory transmitters in the central nervous system, and increase the overall transient excitability, thus triggering seizures (25).
The mutation we found in the Chinese family was localized in the helix-loop-helix structural domain (EF-hand), which is also a Ca2+ binding domain. According to bioinformatics tools (PolyPhen 2 and SIFT), the mutation is predicted to affect protein function (9). Research has shown that human hippocampal neurons carrying the CABP4 c.464G>A (p.G155D) HE mutation had an increased mRNA transcription level but a reduced protein expression level, presumably with abnormal post-translational degradation of amino acid chains or post-translational modifications resulting in unstable CABP4 protein levels. Conversely, the number of dead cells was increased in the mice hippocampal neurons transfected with the mutation, and the electrophysiological functional studies showed a significant increase in the action potential frequency (12). However, there are no subsequent high-quality reports of CABP4-associated epilepsy, and in vitro studies have limitations that do not reflect the effects of CABP4 mutations on intact neural networks, and in general, the epileptogenic mechanism of CABP4 is unclear.
To date, mutations in the CABP4 gene have rarely been reported, and most research has focused on the CABP4 gene in incomplete resting night blindness. Studies have shown that CABP4 is important for synapse structure and functions, such as colocalizing and interacting with expressed voltage-dependent Ca2+ channel Cav1.4 and modulating their functional properties, which should produce a corresponding change in the transmitter release (11). It has also been demonstrated that specific mutations in the CABP4 gene can lead to reduced levels of CABP4 transcripts; however, the remaining CABP4 transcripts may also result in a small number of functional proteins, and as a result, the signaling may not be completely blocked (15).
Epilepsy is a group of disorders, which is mainly characterized by an enduring tendency of generating epileptic seizures, and instantaneous central nervous system dysfunction. The etiology of epilepsies is complicated. According to the 2017 classification of epilepsies by the International League Against Epilepsy, six groups of etiologies need to be considered in clinical practice, including structural, genetic, infectious, metabolic, immune, and unknown etiologies (26). In a retrospective study in Minnesota, about two-thirds of the seizure cases were classified as idiopathic cases, which means that no obvious etiology was found in these cases (27). With the development of sequencing technology, there is increasing evidence that genetic factors play an important role in idiopathic cases (28). More and more genes associated with epilepsy have been reported and identified; however, an enormous number of epileptogenic genes and corresponding epileptogenic mechanisms still need to be explored.
The HE mice with CABP4 mutations did not appear to exhibit abnormal behaviors; however, as the mutation can cause an imbalance in brain protein expression and increased neuronal excitability, it is still of interest to elucidate the relationship between CABP4 mutations and epilepsy. The relationship between CABP4 gene mutations and epilepsy needs to be supported by human genetic data, and further behavioral, and phenotypic studies. With the development of genetic testing tools and advanced research techniques, the exact relationship between the CABP4 gene and epilepsy is expected to be elucidated, and CABP4 might serve as a new target in the diagnosis and treatment of epilepsy.
Conclusions
The results of the study showed that the CABP4 p.G155D mutation increased the expression level of CABP4 in the brainstem and led to a higher discharge frequency of prefrontal cortex neurons, which suggests that the mutation might be involved in seizure onset.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-484/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-484/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-484/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-484/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All the animal-related experimental procedures were performed under a project license (No. 2018003A) granted by the Ethics Committee for Medical Research, Guangdong Provincial People’s Hospital, in compliance with the Chinese National Regulations on the Administration of Laboratory Animals and the Guangdong Provincial People’s Hospital Guideline of Ethical Use and Care of Experimental Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tinuper P, Bisulli F, Cross JH, et al. Definition and diagnostic criteria of sleep-related hypermotor epilepsy. Neurology 2016;86:1834-42. [Crossref] [PubMed]
- Scheffer IE, Bhatia KP, Lopes-Cendes I, et al. Autosomal dominant frontal epilepsy misdiagnosed as sleep disorder. Lancet 1994;343:515-7. [Crossref] [PubMed]
- Steinlein OK, Mulley JC, Propping P, et al. A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 1995;11:201-3. [Crossref] [PubMed]
- De Fusco M, Becchetti A, Patrignani A, et al. The nicotinic receptor beta 2 subunit is mutant in nocturnal frontal lobe epilepsy. Nat Genet 2000;26:275-6. [Crossref] [PubMed]
- Aridon P, Marini C, Di Resta C, et al. Increased sensitivity of the neuronal nicotinic receptor alpha 2 subunit causes familial epilepsy with nocturnal wandering and ictal fear. Am J Hum Genet 2006;79:342-50. [Crossref] [PubMed]
- Dibbens LM, de Vries B, Donatello S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet 2013;45:546-51. [Crossref] [PubMed]
- Heron SE, Smith KR, Bahlo M, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 2012;44:1188-90. [Crossref] [PubMed]
- Combi R, Dalprà L, Ferini-Strambi L, et al. Frontal lobe epilepsy and mutations of the corticotropin-releasing hormone gene. Ann Neurol 2005;58:899-904. [Crossref] [PubMed]
- Chen ZH, Wang C, Zhuo MQ, et al. Exome sequencing identified a novel missense mutation c.464G>A (p.G155D) in Ca2+-binding protein 4 (CABP4) in a Chinese pedigree with autosomal dominant nocturnal frontal lobe epilepsy. Oncotarget 2017;8:78940-7. [Crossref] [PubMed]
- Zeitz C, Kloeckener-Gruissem B, Forster U, et al. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet 2006;79:657-67. [Crossref] [PubMed]
- Haeseleer F, Imanishi Y, Maeda T, et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci 2004;7:1079-87. [Crossref] [PubMed]
- Guo Y, Miao Q, Zhang Y, et al. A novel missense creatine mutant of CaBP4, c.464G>A (p.G155D), associated with autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE), reduces the expression of CaBP4. Transl Pediatr 2022;11:396-402. [Crossref] [PubMed]
- Haeseleer F, Sokal I, Verlinde CL, et al. Five members of a novel Ca(2+)-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem 2000;275:1247-60. [Crossref] [PubMed]
- Cui G, Meyer AC, Calin-Jageman I, et al. Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 channels in mouse auditory hair cells. J Physiol 2007;585:791-803. [Crossref] [PubMed]
- Shaltiel L, Paparizos C, Fenske S, et al. Complex regulation of voltage-dependent activation and inactivation properties of retinal voltage-gated Cav1.4 L-type Ca2+ channels by Ca2+-binding protein 4 (CaBP4). J Biol Chem 2012;287:36312-21. [Crossref] [PubMed]
- Kessi M, Chen B, Peng J, et al. Calcium channelopathies and intellectual disability: a systematic review. Orphanet J Rare Dis 2021;16:219. [Crossref] [PubMed]
- Lauerer RJ, Lerche H. Voltage-gated calcium channels in genetic epilepsies. J Neurochem 2023; Epub ahead of print. [Crossref] [PubMed]
- Faingold CL. The role of the brain stem in generalized epileptic seizures. Metab Brain Dis 1987;2:81-112. [Crossref] [PubMed]
- Kohsaka S, Mizukami S, Uetake K, et al. Brainstem triggers absence seizures in human generalized epilepsy. Brain Res 1999;837:277-88. [Crossref] [PubMed]
- Kohsaka S, Kohsaka M, Mizukami S, et al. Brainstem activates paroxysmal discharge in human generalized epilepsy. Brain Res 2001;903:53-61. [Crossref] [PubMed]
- Papathanasiou ES, Pantzaris M, Myrianthopoulou P, et al. Brainstem lesions may be important in the development of epilepsy in multiple sclerosis patients: an evoked potential study. Clin Neurophysiol 2010;121:2104-10. [Crossref] [PubMed]
- Li HL, Deng ZR, Zhang J, et al. Sonographic hypoechogenicity of brainstem raphe nucleus is correlated with electroencephalographic spike frequency in patients with epilepsy. Epilepsy Behav 2021;117:107884. [Crossref] [PubMed]
- Ishikawa T, Sahara Y, Takahashi T. A single packet of transmitter does not saturate postsynaptic glutamate receptors. Neuron 2002;34:613-21. [Crossref] [PubMed]
- Oertner TG, Sabatini BL, Nimchinsky EA, et al. Facilitation at single synapses probed with optical quantal analysis. Nat Neurosci 2002;5:657-64. [Crossref] [PubMed]
- Zhuo MQ. The electrophysiologic function of CABP4 gene mutation in autosomal dominant norturnal frontal lobe epilepsy. Guangzhou: South Medical University; 2015.
- Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017;58:512-21. [Crossref] [PubMed]
- Hauser WA, Annegers JF, Kurland LT. Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935-1984. Epilepsia 1993;34:453-68. [Crossref] [PubMed]
- Perucca P, Bahlo M, Berkovic SF. The Genetics of Epilepsy. Annu Rev Genomics Hum Genet 2020;21:205-30. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


