
Enhancement of motor neuron development and function in zebrafish by sialyllacto-N-tetraose b
Highlight box
Key findings
• Sialyllacto-N-tetraose b (LSTb) significantly enhances the development and functionality of motor neurons in zebrafish through the upregulation of Slit2 and Slit3.
What is known and what is new?
• Human milk oligosaccharides (HMOs) are widely acknowledged to be essential for brain development and immune system programming, and LSTb is a low-concentration component of HMOs.
• LSTb substantially impacts the development and function of motor neurons in zebrafish.
What is the implication, and what should change now?
• This study’s findings suggest that LSTb may have a broader effect on neurodevelopment, potentially influencing the growth and guidance of neurons beyond the motor system.
• The upregulation of Slit2 and Slit3 indicates a possible mechanism by which LSTb exerts its effects, which could be targeted for therapeutic interventions in neurodevelopmental disorders.
• Further research should focus on the effects of LSTb in other animal models and eventually in human clinical trials to explore its potential as a therapeutic agent for neurodevelopmental disorders.
• The findings also underscore the importance of including LSTb in the formulation of infant formulas to potentially mimic the benefits of breast milk and support optimal brain development.
Introduction
Human milk oligosaccharides (HMOs) are a type of polysaccharide and the third largest component of human milk after lactose and lipids (1). They serve as prebiotics and play a significant role in brain development and in programming the immune system to reduce the incidence of allergies and autoimmune diseases (2,3). More than 200 different structures of HMOs have been identified in human milk. They primarily consist of monosaccharides such as D-glucose (Glc), D-galactose (Gal), N-acetyl-D-glucosamine (GlcNAc), L-fucose (Fuc), and N-acetylneuraminic acid (Neu5Ac) (4), which form a series of complex oligosaccharides that can be classified into three major types based on their fucosylation or sialylation: (I) neutral nonfucosylated HMOs; (II) neutral fucosylated HMOs; and (III) HMOs containing sialic acid (5,6).
Sialyllacto-N-tetraose b (LSTb) is a type of sialylated oligosaccharide found in mammal milk. It comprises three monosaccharide units, including galactose, glucose, and lactose. There are individual differences in the concentrations and distributions of LSTb in breast milk (7,8). However, due to the low concentration of LSTb in milk, research on its effects is limited. Whether LSTb, similar to other HMOs, affects the development of the central nervous system is still poorly understood.
Zebrafish have gained recognition as a popular model organism in neurobiology in recent years (9). Both adult and larval zebrafish have greatly contributed to neuroscience due to their remarkable physiological and genetic similarities to humans (10,11), as well as their amenability to gene manipulation and comparable central nervous system morphology (12). Zebrafish provide a stable platform that balances the intricacy of a vertebrate and its utility as a model organism. With the added benefits of optical transparency in developing fish facilitating the application of advanced imaging techniques, these complex mechanisms at the organism level can be effectively visualized (13).
This study used the transgenic (Tg) zebrafish line (Hb9:GFP), which is commonly used to evaluate motor neurons of the central nervous system. We incubated the zebrafish with LSTb to gain insight into its influence on nervous system development. We present this article in accordance with the MDAR and ARRIVE reporting checklists (available at https://tp.amegroups.com/article/view/10.21037/tp-24-247/rc).
Methods
Zebrafish husbandry
Specimens of the Tg zebrafish line (Hb9:GFP) were acquired from the Zebrafish Facility of Nantong University. Experiments were performed under a project license (No. S20210310-007) granted by the ethics committee of Nantong University, in compliance with institutional guidelines for the care and use of animals. A protocol was prepared before the study without registration. The zebrafish larvae were kept in a recirculation system under a 14-hour light and 10-hour dark photocycle at 28±0.5 ℃ and a pH range of 7.0–7.5. Embryos at 8 hours post-fertilization (hpf) were co-incubated with 1 mg/mL of LSTb (DSM, Maastricht, the Netherlands) as the LSTb group. The control group was incubated according to the normal process.
Image observation of Tg zebrafish
Zebrafish larvae were placed in 0.7% low-melting-point agarose. Confocal imaging was carried out at 72 hpf. Embryos were anesthetized with egg water and 0.16 mg/mL of tricaine to acquire good pictures. Confocal imaging was accomplished using the TCS-SP5 laser scanning microscope (Leica, Wetzlar, Germany) and analyzed using Imaris (Oxfords Instruments, Abingdon, UK) and ImageJ (National Institutes of Health, Bethesda, MD, USA) software.
Zebrafish larva locomotor behavior test
In accordance with previously established methods (14), 30 zebrafish larvae at 5 days post-fertilization (dpf) were selected at random from groups and plated individually within each well of a 24-well plate. The DanioVision Observation Chamber (Noldus, Wageningen, the Netherlands) was employed to facilitate the measurement and observation of the swimming behavior of the larvae. Prior to monitoring, the larvae were allowed to acclimate for a period of 30 min at 28 ℃. The Ethovision XT 13 system (Noldus) was then used to complete the locomotor behavior test of the zebrafish larvae within 30 min. The software computed the locomotor behavior distance and spontaneous movement, with the average speed being derived for each group.
RNA sequencing (RNA-seq) analysis
We performed RNA-seq at 72 hpf. Total RNA from zebrafish was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and reverse-transcribed into complement DNA (cDNA) using the first strand of transcriptional cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany). Library sequencing was performed using a HiSeq 4000 platform (Illumina, San Diego, CA, USA). Raw readings were filtered to exclude low-quality data, and high-quality raw data were used for downstream analyses. Differential expression analysis was completed with the DESeq2 (v. 1.6.3) R package. Differential gene expression changes of more than two-fold and q value ≤0.05 indicated a significant difference. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the differentially expressed genes (DEGs) were completed.
Real-time polymerase chain reaction (real-time PCR)
Total RNA was extracted using TRIzol UP (TransGen Biotech, Beijing, China) and transcribed into cDNA using a HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The primer used was Slit2 and Slit3 (Slit2 forward primer: GCTGTTCAGGCGTAGAAGACTACC; Slit2 reverse primer: GTATACTGAGGGATGTGGTCTGGGA; Slit3 forward primer: TCCCAACTTGTCCTACCTGTAGTGT; Slit3 reverse primer: ACCCACCGCATCATATATCCTCTGA).
Whole-mount in situ hybridization
The cDNA served as a template for cloning fragments of Slit2 to create antisense RNA probes for zebrafish Slit2 and Slit3 at 24, 48, and 72 hpf. The Slit2 primers were determined via reverse transcription PCR (RT-PCR). Digoxigenin (DIG)-labeled RNA sense and antisense probes were generated from the linearized plasmids with a DIG RNA Labeling Kit (SP6/T7) (Roche Diagnostics). Whole-mount in situ hybridization was conducted as per previously reported procedures (15).
Measurement and statistical analysis
The axonal number and length of caudal primary (CaP) neurons were measured using ImageJ software. The branch numbers within the distal 50 µm and axonal length were calculated. All data analysis and statistical comparisons were completed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). Statistical significance was determined using the Student’s t-test. Data are presented as the mean ± standard deviation, and P values <0.05 were considered to be statistically significant.
Results
Effects of LSTb on the axonal growth of CaP neurons
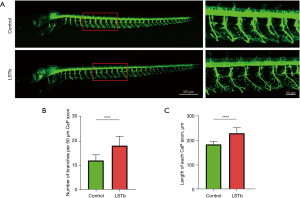
LSTb treatment significantly increased the number of CaP axon branches per 50 µm at 72 hpf compared with that observed in the control group (18.05±0.8568 vs. 12.00±0.5282) (Figure 1A,1B). The average length of the CaP axon in the LSTb group (229.4±4.219 µm) was significantly longer than that of the control group (183.9±2.119 µm) (Figure 1A,1C).
Locomotor behavioral changes induced by LSTb
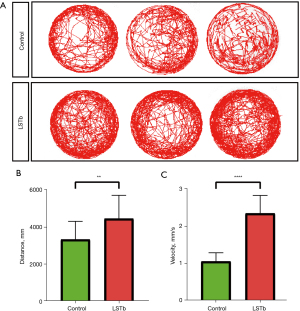
As shown in Figure 2A, zebrafish larvae of the LSTb group exhibited more frequent behavioral trajectories at 5 dpf. In the LSTb group, the distance and speed of the zebrafish larvae notably increased (Figure 2B,2C).
Transcriptomic profiling of LSTb treatment
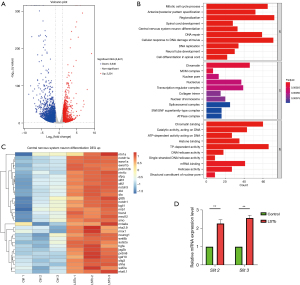
We identified 5,847 DEGs between the two groups, with 2,201 genes being upregulated and 3,646 genes being downregulated (Figure 3A). According to GO annotations, central nervous system neuron differentiation was noted as the representative GO term enriched from DEGs in biological processes (BP) (Figure 3B). We also observed 33 upregulated DEGs in GO term of central nervous system neuron differentiation. Among these 33 DEGs, Slit2 and Slit3 are classic genes for axon guidance (Figure 3C). To verify the reliability of RNA-seq, we conducted quantitative RT-PCR on the expression of Slit2 and Slit3 at 72 hpf. The expression of Slit2 and Slit3 was consistent with the that from the RNA-seq results, with significantly increased expression in the LSTb group (Figure 3D).
Expression of Slit2 in zebrafish
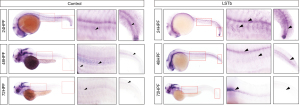
We analyzed the temporal and spatial expression patterns of Slit2 in embryos at 24, 48, and 72 hpf. Slit2 was ubiquitously expressed in the central nervous system, including the brain and spinal cord. Notably, Slit2 expression was more enhanced in the LSTb group compared with that in the control group after fertilization (Figure 4). This suggested that LSTb could promote the expression of Slit2 in the central nervous system.
Discussion
Sialylated HMOs (SHMOs) are a type of HMO that constitute about 20% of all HMOs (16,17). SHMOs play an important role in the growth and development of the brain and nervous system in infants (18,19) and serve as a source of nutrients for infant neurodevelopment. LSTb is a specific type of SHMO (molecular formula: C37H62N2O29; molecular weight: 998.9 g/mol) and consists of one lactosamine Galβ1-3/4GlcNAc, one lactose core Galβ1-4Glc, and one Neu5Ac. The structural sequence of LSTb is Galβ1-3 (Neu5Acα2-6) GlcNAcβ1-3Galβ1-4Glc (Neu5Ac 1 Gal 2 Glc 2).
The levels of LSTb in human breast milk are relatively low and can be affected by various factors. This includes regional differences, as shown by the modest LSTb level of 41±5 nmol/mL seen in Peruvian women (7). LSTb concentrations typically peak during lactation days 8 to 10 and then decrease (10.3390/nu15061408). Additionally, a negative correlation was found between maternal body mass index at 5 months and LSTb level (20). A study on the relationship between LSTb and infants revealed that LSTb levels were negatively associated with weight and head growth at 14 and 28 days postpartum (21). LSTb levels at 6 months were also demonstrated to be predictive of cognitive development scores in infants (22). It is possible that LSTb levels could be linked to infant cognitive development through the proliferation or depletion of specific gut microbes (23,24).
The impact of LSTb on cognitive performance in infants differs from that of other HMO constituents. To investigate the effects of LSTb on central nervous system development, we opted to use Tg zebrafish and study motor neuron development. The results indicated that coincubation of zebrafish with LSTb increased both the number and length of CaP neuron branches, suggesting that LSTb indeed facilitates the growth of CaP neurons. CaP, a type of primary motor neuron, is closely related to locomotion. Thus, we conducted behavioral testing, which showed that immersion of zebrafish larvae in LSTb resulted in notable enhancements in swimming distance, tail-wagging movement, and average speed. RNA-seq was performed to understand how LSTb promotes CaP development, and the results showed that zebrafish larvae soaked in LSTb exhibited increased expression in 33 genes, with GO entries enriched in central nervous system neuron differentiation. Slit2 and Slit3, classic genes related to axon guidance became the focus of our attention. Subsequent verification by PCR also showed that the expression of Slit2 and Slit3 increased significantly after LSTb treatment. The whole-mount in situ hybridization results also showed that the expression of Slit2 at different times after incubation with LSTb was stronger than that in the control group. Based on these findings, we speculate that LSTb promotes CaP neuron development through Slit2 and Slit3.
The Slit family is a group of secreted proteins that hold critical significance in guiding neuronal axons. Slit proteins consist of single peptides that contain about 1,500 amino acids. In vertebrates, the Slit family includes Slit1, Slit2, and Slit3 (25). In zebrafish, four genes have been characterized, namely Slit1a, Slit1b, Slit2, and Slit3 (26,27). Among the Slit proteins, Slit1 is mainly expressed within the developing nervous system, while Slit2 and Slit3 are present in other parts of the body, such as the kidney, lungs, heart and, immune cells, in addition to the nervous system (28-30). A study conducted in the late 1990s showed that Slit2 within the calf brain led to a fivefold increase in the number of branch points per axon and to 2.5-fold increase in axon length (31). Slit has been identified as a positive regulator of dorsal root ganglion axonal branching and elongation (32). Slits typically exert their biological function by binding to roundabout (Robo) receptors, and Slit-Robo signaling plays a role in various BP, including axon guidance and central nervous system development (33).
There are some limitations to this study that should be mentioned. Although the study examined the changes of structure of CaP neurons and motor ability, the effect of LSTb on motor neuron differentiation was not observed, and other types of neurons in the central nervous system and their corresponding functions were not taken into account. Additionally, we used zebrafish and did not investigate the higher functions of the central nervous system, such as learning and memory. Our findings indicated that LSTb, despite not being a major component of breast milk, can significantly impact the development of motor neurons. The mechanism behind this effect may be related to the increase in the expression of Slit2 and Slit3 by LSTb. In order to further explore the impact of LSTb on the central nervous system, we plan to conduct more research using mammalian models in the future, providing more reliable experimental data and theoretical support for the addition of LSTb to infant formula.
Conclusions
The study provides evidence that LSTb, despite its low concentration in human milk, has a significant impact on the development and function of motor neurons in zebrafish. LSTb may modulate neurodevelopment by regulating the Slit2 and Slit3 genes that are critical for axonal growth and guidance. These findings contribute to our understanding of the role of HMOs in neurodevelopment and provide new avenues for further exploration of LSTb’s potential as a therapeutic agent for neurodevelopmental disorders.
Acknowledgments
Funding: This research was funded by
Footnote
Reporting Checklist: The authors have completed the MDAR and ARRIVE reporting checklists. Available at https://tp.amegroups.com/article/view/10.21037/tp-24-247/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-24-247/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-24-247/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-24-247/coif). All authors report that this research was funded by the Key Medical Research Project of Jiangsu Provincial Health Commission (No. ZD2021004), the Maternal and Child Health Research Project of Jiangsu Province (Nos. FYX202125 and F202330), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. SJCX23_1798), the Science and Technology Program of Nantong (Nos. JC12022038 and JC22022028), and the Project of Nantong Health Commission (Nos. QN2022006 and QB2021002). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. S20210310-007) granted by the ethics committee of Nantong University, in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Austin S, De Castro CA, Bénet T, et al. Temporal Change of the Content of 10 Oligosaccharides in the Milk of Chinese Urban Mothers. Nutrients 2016;8:346. [Crossref] [PubMed]
- Plaza-Díaz J, Fontana L, Gil A. Human Milk Oligosaccharides and Immune System Development. Nutrients 2018;10:1038. [Crossref] [PubMed]
- Dinleyici M, Barbieur J, Dinleyici EC, et al. Functional effects of human milk oligosaccharides (HMOs). Gut Microbes 2023;15:2186115. [Crossref] [PubMed]
- Vandenplas Y, Berger B, Carnielli VP, et al. Human Milk Oligosaccharides: 2'-Fucosyllactose (2'-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula. Nutrients 2018;10:1161. [Crossref] [PubMed]
- Smilowitz JT, Lebrilla CB, Mills DA, et al. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr 2014;34:143-69. [Crossref] [PubMed]
- Van Niekerk E, Autran CA, Nel DG, et al. Human milk oligosaccharides differ between HIV-infected and HIV-uninfected mothers and are related to necrotizing enterocolitis incidence in their preterm very-low-birth-weight infants. J Nutr 2014;144:1227-33. [Crossref] [PubMed]
- McGuire MK, Meehan CL, McGuire MA, et al. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 2017;105:1086-100. [Crossref] [PubMed]
- Ren X, Yan J, Bi Y, et al. Human Milk Oligosaccharides Are Associated with Lactation Stage and Lewis Phenotype in a Chinese Population. Nutrients 2023;15:1408. [Crossref] [PubMed]
- Adams MM, Kafaligonul H. Zebrafish-A Model Organism for Studying the Neurobiological Mechanisms Underlying Cognitive Brain Aging and Use of Potential Interventions. Front Cell Dev Biol 2018;6:135. [Crossref] [PubMed]
- Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013;496:498-503. [Crossref] [PubMed]
- Shehwana H, Konu O. Comparative Transcriptomics Between Zebrafish and Mammals: A Roadmap for Discovery of Conserved and Unique Signaling Pathways in Physiology and Disease. Front Cell Dev Biol 2019;7:5. [Crossref] [PubMed]
- Kalueff AV, Stewart AM, Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol Sci 2014;35:63-75. [Crossref] [PubMed]
- Saleem S, Kannan RR. Zebrafish: A Promising Real-Time Model System for Nanotechnology-Mediated Neurospecific Drug Delivery. Nanoscale Res Lett 2021;16:135. [Crossref] [PubMed]
- Gu J, Zhang J, Chen Y, et al. Neurobehavioral effects of bisphenol S exposure in early life stages of zebrafish larvae (Danio rerio). Chemosphere 2019;217:629-35. [Crossref] [PubMed]
- Dong Z, Chen X, Li Y, et al. Microtubule Severing Protein Fignl2 Contributes to Endothelial and Neuronal Branching in Zebrafish Development. Front Cell Dev Biol 2020;8:593234. [Crossref] [PubMed]
- Ninonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. J Agric Food Chem 2006;54:7471-80. [Crossref] [PubMed]
- Bode L, Kunz C, Muhly-Reinholz M, et al. Inhibition of monocyte, lymphocyte, and neutrophil adhesion to endothelial cells by human milk oligosaccharides. Thromb Haemost 2004;92:1402-10. [Crossref] [PubMed]
- Varki A. Sialic acids in human health and disease. Trends Mol Med 2008;14:351-60. [Crossref] [PubMed]
- Wu S, Grimm R, German JB, et al. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res 2011;10:856-68. [Crossref] [PubMed]
- Larsson MW, Lind MV, Laursen RP, et al. Human Milk Oligosaccharide Composition Is Associated With Excessive Weight Gain During Exclusive Breastfeeding-An Explorative Study. Front Pediatr 2019;7:297. [Crossref] [PubMed]
- Wejryd E, Martí M, Marchini G, et al. Low Diversity of Human Milk Oligosaccharides is Associated with Necrotising Enterocolitis in Extremely Low Birth Weight Infants. Nutrients 2018;10:1556. [Crossref] [PubMed]
- Mulinge MM, Abisi HK, Kabahweza HM, et al. The Role of Maternal Secretor Status and Human Milk Oligosaccharides on Early Childhood Development: A Systematic Review and Meta-Analysis. Breastfeed Med 2024;19:409-24. [Crossref] [PubMed]
- Moossavi S, Atakora F, Miliku K, et al. Integrated Analysis of Human Milk Microbiota With Oligosaccharides and Fatty Acids in the CHILD Cohort. Front Nutr 2019;6:58. [Crossref] [PubMed]
- Berger PK, Plows JF, Jones RB, et al. Human milk oligosaccharide 2'-fucosyllactose links feedings at 1 month to cognitive development at 24 months in infants of normal and overweight mothers. PLoS One 2020;15:e0228323. [Crossref] [PubMed]
- Dickson BJ, Gilestro GF. Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu Rev Cell Dev Biol 2006;22:651-75. [Crossref] [PubMed]
- Yeo SY, Little MH, Yamada T, et al. Overexpression of a slit homologue impairs convergent extension of the mesoderm and causes cyclopia in embryonic zebrafish. Dev Biol 2001;230:1-17. [Crossref] [PubMed]
- Hutson LD, Jurynec MJ, Yeo SY, et al. Two divergent slit1 genes in zebrafish. Dev Dyn 2003;228:358-69. [Crossref] [PubMed]
- Wu JY, Feng L, Park HT, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature 2001;410:948-52. [Crossref] [PubMed]
- Prasad A, Qamri Z, Wu J, et al. Slit-2/Robo-1 modulates the CXCL12/CXCR4-induced chemotaxis of T cells. J Leukoc Biol 2007;82:465-76. [Crossref] [PubMed]
- Chaturvedi S, Robinson LA. Slit2-Robo signaling in inflammation and kidney injury. Pediatr Nephrol 2015;30:561-6. [Crossref] [PubMed]
- Zinn K, Sun Q. Slit branches out: a secreted protein mediates both attractive and repulsive axon guidance. Cell 1999;97:1-4. [Crossref] [PubMed]
- Wang KH, Brose K, Arnott D, et al. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell 1999;96:771-84. [Crossref] [PubMed]
- Sherchan P, Travis ZD, Tang J, et al. The potential of Slit2 as a therapeutic target for central nervous system disorders. Expert Opin Ther Targets 2020;24:805-18. [Crossref] [PubMed]


