
Association of human adenovirus load and viral genotype diversity with respiratory disease severity in children: a systematic review and meta-analysis
Highlight box
Key findings
• In children, there was a significant association between the load of human adenovirus (HAdV) subtypes B3 and B7 and respiratory disease severity. Quantitative polymerase chain reaction demonstrated high sensitivity and specificity for detecting the B3 and B7 virus subtypes, suggesting its suitability for clinical use.
What is known and what is new?
• Acute respiratory diseases are the leading cause of hospital admissions in children. Among these, HAdVs remain one of the most prevalent viruses in pediatric outpatients worldwide despite the relatively low rate of hospital admissions in these cases. HAdV infections are widely distributed in nature and have been reported in various countries and regions.
• The correlation between different genotypes and disease severity requires further investigation. This meta-analysis assessed the association among HAdV load, genotypic diversity, and the severity of respiratory disease in children.
What is the implication, and what should change now?
• This study provides valuable insights into the role of HAdV in childhood respiratory infections. Future studies could use molecular biology and immunological approaches to investigate how HAdV affects host immune responses and how these responses influence disease severity.
Introduction
Acute respiratory diseases are the leading cause of hospital admissions in children. Among these, human adenoviruses (HAdVs) remain one of the most common viruses in pediatric outpatients worldwide (1) despite the relatively low rate of hospital admissions in patients with this disease (2). HAdV infections are widely distributed in nature and have been reported in various countries and regions (3). They account for a significant proportion of hospital admissions for pneumonia, acute bronchiolitis, and viral respiratory infection in children (4,5). A study in Argentina found that the mortality rate of children with adenoviral acute lower respiratory tract infection was as high as 16.7% (6). Of these cases, 71% were associated with pneumonia, and 86.4% of HAdV-infected children were younger than 5 years (7), suggesting that adenovirus (AdV) infection is a serious threat. Furthermore, a subset of pediatric patients infected with HAdV may develop severe chronic complications, notably post-infectious bronchiolitis obliterans (PIBO) and hemophagocytic syndrome, which represent potentially life-threatening sequelae (8). Timely and accurate assessment and prevention of AdV infection can improve the treatment of HAdV-associated respiratory diseases.
As methods for measuring viral load in respiratory samples have become standardized (9), the viral load of HAdVs has become an important indicator for assessing infection severity in clinical practice. Children with severe AdV pneumonia, especially those with prolonged fever (≥10.5 days) who develop respiratory distress or require invasive mechanical ventilation during the acute phase, are more likely to develop bronchiolitis obliterans (10). Specific genotypes such as HAdV-B3 are associated with increased viral load in respiratory samples and correlate with clinical presentation and outcome (11), with HAdV-1 and HAdV-31 strongly correlated with greater disease severity in children (12). The persistence of viral shedding in children with severe HAdV pneumonia varies by genotype; for instance, viral shedding of HAdV-7 may persist for more than 3 months and decline more slowly than that of HAdV-3. These genotype-specific variations in viral dynamics can inform clinical management strategies and enhance prognostic evaluations (13). Therefore, monitoring viral load kinetics in HAdV pneumonia, especially in severely ill children, is valuable for understanding the disease process, guiding clinical treatment strategies, and evaluating efficacy.
In China, a study of 1,447 children revealed that HAdV-3 and HAdV-7 are the dominant subtypes in childhood influenza-like illnesses (14). The major pathogenic targets of HAdVs include processes such as the cell cycle, mitosis, DNA replication, and intracellular transport of the virus (15). In cases of co-infection with other viruses, HAdV infection primarily presents with fever, cough, and sputum production. Specific genotypes, such as HAdV-B3, may influence disease severity (16). The prevalence trends and pathogenic potential of these genotypes vary by region, suggesting the need for region-specific studies of HAdV genotype diversity. New pathogenic genomic variants, such as HAdV-7, have been identified and associated with severe sequelae and high mortality (17). A study in South America has demonstrated an association between severe respiratory infections and subspecies B1 (18). Additionally, the high serum viral load associated with a severe cytokine storm following HAdV-7 infection may significantly contribute to the poor prognosis in children (19). However, the correlation between different genotypes and disease severity requires further investigation.
This meta-analysis assessed the association among HAdV load, genotypic diversity, and the severity of respiratory disease in children. We conducted a systematic review and quantitative analysis of the literature to address inconsistent findings and explore the role of viral load and the genotypic diversity of HAdVs in childhood respiratory diseases. We hypothesize that specific AdV genotypes are associated with the severity of respiratory disease in children and that HAdV viral load can predict disease severity. We expect this study to provide a more accurate clinical predictor and scientific basis for preventing and treating HAdV infections in children. We present this article in accordance with the MOOSE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-2024-627/rc).
Methods
Literature search
To comprehensively evaluate the association among HAdV load, viral genotypic diversity, and severity of respiratory disease in children, we conducted a systematic search of Web of Science, PubMed, American Medical Association (AMA), Cochrane Library, China National Knowledge Infrastructure (CNKI), and European Society for Clinical Nutrition and Metabolism (ESPEN) databases to compare relevant studies on the association between respiratory AdV infections of different severity and viral load and genotyping. The main search terms included “adenovirus”, “pneumonia”, “bronchitis”, “viral load”, and “genotyping”, among others. The language of the literature was limited to English or Chinese. The selection of databases was based on their comprehensive coverage of biomedical literature (Web of Science, PubMed, AMA, Cochrane Library) and regional relevance (CNKI for Chinese studies, ESPEN for European context), while study inclusion prioritized alignment with research objectives through keywords addressing disease severity, viral load, and genotyping to ensure representativeness and minimize language bias.
Literature inclusion criteria
The inclusion criteria for the literature were: (I) observational study; (II) comparison of the relationship between different levels of severity and AdV viral load and/or genotyping in children; (III) reporting of AdV viral load, disease severity, and genotyping-related indicators; and (IV) provision of reports of effect indicators or original data from which the corresponding indicators could be calculated.
Exclusion criteria for literature
The exclusion criteria for the literature were: (I) review studies, syntheses, or case reports; (II) no reporting of outcome indicators of respiratory disease severity; and (III) duplicate publications or duplicate data.
Data collection
Two reviewers independently searched and screened the literature to determine the study eligibility based on the inclusion and exclusion criteria. Disagreements were resolved by discussion and consensus. After data extraction, meta-analysis was performed using RevMan 5.3 software (Cochrane, London, UK) to assess the association among disease severity, HAdV load, and genotyping in children.
Quality assessment of the literature
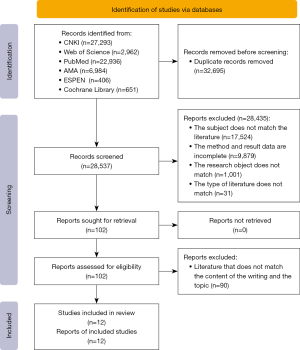
Information including study type, sample size, effect indicator classification, and data were extracted from the included studies. Two researchers, R.Z. and X.Z., extracted the information independently, checked it for consistency, and entered it into the statistical software. The Cochrane Rating Tool was used to assess the risk of bias in the included trials. The assessment criteria were as follows: grade A, low risk of bias; grade B, moderate risk of bias; and grade C, high risk of bias. The process and results of the literature search are detailed in Figure 1.
Statistical analyses
RevMan 5.3 software and STATA version 15.1 (StataCorp LLC, College Station, TX, USA) were used for the statistical analyses. Viral load was examined using odds ratio (OR) and standard error (SE) as effect sizes, and the association between viral genotyping and children with severe pneumonia was examined using true positive, false positive, true negative, and false negative as effect sizes. Heterogeneity between studies was assessed, with I2=0 indicating no statistical heterogeneity, I2<50% indicating moderate heterogeneity, and I2>50% indicating greater heterogeneity. Funnel plots and Deeks’ method were used to assess the presence of publication bias. Subgroup analyses were performed to assess the robustness of study results according to the study sample size and test method.
Results
Characteristics of the included studies
Twelve studies (13,19-29) published from 2015 to 2024 were ultimately retrieved. The studies covered five countries: China, Israel, Germany, Italy, and Vietnam. Three studies used nested polymerase chain reaction (PCR), eight used real-time fluorescence-based quantitative PCR (qPCR), and one used real-time PCR. The total sample size was 2,194 cases, of which 554 were severe. The relevant indices included four articles on viral load; six articles with a B3 viral type; eight articles with a B7 viral type; and two articles with the C1, C2, and C5 subtypes. The characteristics of each study and other relevant indices are shown in Table 1.
Table 1
| Author | Year | Country | Detection method | Sample size/number of severe pneumonia (%) | Relevant indicators |
|---|---|---|---|---|---|
| Zeng et al. (13) | 2021 | China | Fluorescence-based quantitative PCR | 117/87 (74.3) | B3, B7 |
| Wei et al. (19) | 2023 | China | Nested PCR products | 120/37 (30.8) | B3, B7, C1, C2, C5, C6, C2/6, C57, D37 |
| Esposito et al. (20) | 2016 | Italy | Real-time PCR | 59/10 (16.9) | Viral load |
| Fu et al. (21) | 2019 | China | Quantitative PCR | 158/62 (39.2) | B3, B7, |
| Goikhman et al. (22) | 2020 | Israel | Quantitative PCR | 123/75 (60.9) | Viral load |
| Lin et al. (23) | 2017 | China | Fluorescence-based quantitative PCR | 621/75 (12.1) | B2, B3, B7 |
| Nguyen et al. (24) | 2023 | Vietnam | Nested PCR products | 29/5 (17.2) | B7, B8 |
| Papan et al. (25) | 2023 | Germany, Italy | Fluorescence-based quantitative PCR | 333/13 (3.9) | Viral load |
| Wang et al. (26) | 2024 | China | Nested PCR products | 121/15 (12.4) | B3, B4, B7, undivided |
| Wo et al. (27) | 2015 | China | Fluorescence-based quantitative PCR | 208/84 (40.3) | B3, B7 |
| Xie et al. (28) | 2021 | China | Fluorescence-based quantitative PCR | 94/31 (32.9) | B7 |
| Zhang et al. (29) | 2021 | China | Fluorescence-based quantitative PCR | 111/60 (54.1) | Viral load |
PCR, polymerase chain reaction.
Quality evaluation of the included literature
As shown in Figure 2, most of the 12 articles were low risk and high quality, meeting the requirements for further analyses.
Comparison of methodological results
Association between viral load and severe pneumonia
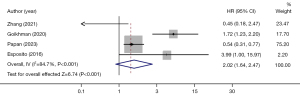
Of the 12 papers, 4 examined viral load, including 158 cases of severe pneumonia, for a total sample size of 626 cases (Figure 3). The I2 was 84.7%, indicating high heterogeneity in the literature, with a combined OR of 2.02 [95% confidence interval (CI): 1.64–2.47]. The risk of developing severe pneumonia among groups with variable viral load was statistically significant (Z=6.74; P<0.001). A high viral load increased the risk of severe pneumonia.
Association of B3 subtype with severe pneumonia
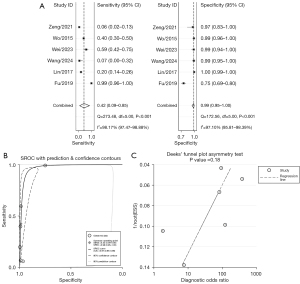
Among the infected cases, 360 had severe pneumonia. As determined with Stata/SE 15.1 software, the combined effect sizes were as follows: the pooled sensitivity was 0.42 (95% CI: 0.09–0.85), the pooled specificity was 0.99 (95% CI: 0.95–1.00), the pooled positive likelihood ratio was 34.4 (95% CI: 15.8–74.8), the pooled negative likelihood ratio was 0.58 (95% CI: 0.25–1.35), and the pooled diagnostic OR was 59 (95% CI: 20–170). An asymmetry test (Deeks’ method) yielded a P value of 0.18, suggesting no significant evidence of publication bias. The area under the pooled receiver operating characteristic (ROC) curve was 0.97 (95% CI: 0.95–0.98). See Figure 4 for more details.
Comparison of association between the B7 subtype and severe pneumonia
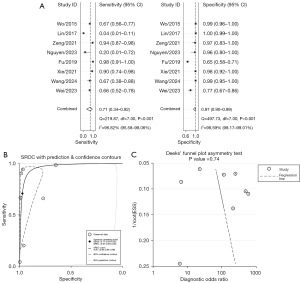
Of the 12 studies on AdV infection, 8 were related to subtype B7 and included 1,568 children with lung infections, 396 of whom had severe pneumonia. As shown in Figure 5, the sensitivity estimates from each study were relatively dispersed, while the specificity estimates were more clustered. The combined effect sizes, calculated using Stata/SE 15.1 software, were as follows: the pooled sensitivity was 0.71 (95% CI: 0.34–0.92), the pooled specificity was 0.97 (95% CI: 0.90–0.99), the pooled positive likelihood ratio was 26.6 (95% CI: 8.0–88.1), the pooled negative likelihood ratio was 0.30 (95% CI: 0.10–0.88), and the pooled diagnostic OR was 89 (95% CI: 23–344). The area under the combined ROC curve was 0.96 (95% CI: 0.94–0.98). An asymmetry test (Deeks’ method) yielded a P value of 0.74, suggesting no evidence of substantial publication bias. See Figure 5 for more details.
Subgroup analyses
Subgroup analysis of viral load
For sample sizes ≤100, the combined OR was 0.88 (95% CI: 0.41–1.90). For sizes >100, the OR was 2.15 (95% CI: 1.74–2.65). When the sample size increased to ≤200, the OR was 3.29 (95% CI: 2.18–4.95), and for >200, it was 1.72 (95% CI: 1.36–2.17). This shows that the combined OR increased with the increase in sample size. However, all CIs included a value of 1, indicating no significant association. The OR for real-time PCR was 0.45 (95% CI: 0.18–1.14). With qPCR, the combined OR increased to 2.18 (95% CI: 1.77–2.69), suggesting that the viral load determined by qPCR had more clinical value. For more details, refer to Table 2.
Table 2
| Subgroup | Authors | OR (95% CI) |
|---|---|---|
| Sample size | ||
| ≤100 | Zhang, Esposito | 0.88 (0.41–1.90) |
| >100 | Goikhma, Papan | 2.15 (1.74–2.65) |
| ≤200 | Zhang, Goikhman, Esposito | 3.29 (2.18–4.95) |
| >200 | Papan | 1.72 (1.36–2.17) |
| Real-time PCR | Esposito | 0.45 (0.18–1.14) |
| Quantify PCR | Zhang, Goikhman, Papan | 2.18 (1.77–2.69) |
CI, confidence interval; OR, odds ratio; PCR, polymerase chain reaction.
Subgroup analysis of viral genotyping
Subtype B3 viruses: for sample sizes ≤150, the combined sensitivity was 0.20 (95% CI: 0.13–0.28), and the combined specificity was 0.96 (95% CI: 0.95–0.97). For sample sizes >150, the combined sensitivity increased to 0.48 (95% CI: 0.43–0.53), while the combined specificity was 0.92 (95% CI: 0.85–0.98). For sample sizes ≤200, the combined sensitivity was 0.58 (95% CI: 0.55–0.60), and the combined specificity was 0.86 (95% CI: 0.85–0.87). For sample sizes >200, the combined sensitivity decreased to 0.26 (95% CI: 0.21–0.31), but the combined specificity increased to 0.99 (95% CI: 0.99–1.00). These results suggest a significant association between sample size and test performance, as the CI values did not include 1. In the comparison of assay methods, qPCR showed a combined sensitivity of 0.48 (95% CI: 0.43–0.53) and combined specificity of 0.92 (95% CI: 0.85–0.98). By contrast, nested PCR had a combined sensitivity of 0.20 (95% CI: 0.13–0.28) and combined specificity of 0.96 (95% CI: 0.95–0.97). These findings suggest that real-time qPCR assays for B3 subtype viruses have superior clinical value.
Subtype B7 viruses: for sample sizes ≤120, the combined sensitivity was 0.80 (95% CI: 0.49–0.94), and the combined specificity was 0.95 (95% CI: 0.81–0.99). For sample sizes >120, the combined sensitivity was 0.63 (95% CI: 0.13–0.95), and the combined specificity was 0.98 (95% CI: 0.82–1.00). For sample sizes ≤200, the combined sensitivity was 0.84 (95% CI: 0.56–0.95), and the combined specificity was 0.94 (95% CI: 0.81–0.99). For sample sizes >200, the combined sensitivity was 0.37 (95% CI: 0.29–0.45), and the combined specificity was 0.99 (95% CI: 0.99–1.00). As sample size increased, the combined sensitivity and specificity improved, with no CI values of 1, indicating a significant correlation. Real-time qPCR yielded a combined sensitivity of 0.79 (95% CI: 0.29–0.97) and a specificity of 0.98 (95% CI: 0.85–1.00), while nested PCR showed a combined sensitivity of 0.63 (95% CI: 0.52–0.74) and specificity of 0.91 (95% CI: 0.85–0.97). These results suggest that real-time qPCR for the B7 subtype viruses have superior clinical value. See Table 3 for the detailed results.
Table 3
| Viral type | Subgroup classification | Authors | Combined sensitivity (95% CI) | Combined specificity (95% CI) |
|---|---|---|---|---|
| Subtype B3 | ≤150 | Wei, Wang, Zeng | 0.20 (0.13–0.28) | 0.96 (0.95–0.97) |
| >150 | Fu, Lin, Wo | 0.48 (0.43–0.53) | 0.92 (0.85–0.98) | |
| ≤200 | Wei, Wang, Zeng, Fu | 0.58 (0.55–0.60) | 0.86 (0.85–0.87) | |
| >200 | Lin, Wo | 0.26 (0.21–0.31) | 0.99 (0.99–1.00) | |
| Fluorescence-based qPCR | Fu, Lin, Wo | 0.48 (0.43–0.53) | 0.92 (0.85–0.98) | |
| Nested PCR products | Wei, Wang, Zeng | 0.20 (0.13–0.28) | 0.96 (0.95–0.97) | |
| Subtype B7 | ≤120 | Wei, Xie, Nguyen, Zeng | 0.80 (0.49–0.94) | 0.95 (0.81–0.99) |
| >120 | Wang, Fu, Lin, Wo | 0.63 (0.13–0.95) | 0.98 (0.82–1.00) | |
| ≤200 | Wei, Wang, Xie, Nguyen, Zeng, Fu | 0.84 (0.56–0.95) | 0.94 (0.81–0.99) | |
| >200 | Lin, Wo | 0.37 (0.29–0.45) | 0.99 (0.99–1.00) | |
| Fluorescence-based qPCR | Fu, Xie, Zeng, Lin, Wo | 0.79 (0.29–0.97) | 0.98 (0.85–1.00) | |
| Nested PCR products | Wei, Wang, Nguyen | 0.63 (0.52–0.74) | 0.91 (0.85–0.97) |
CI, confidence interval; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
Discussion
Early detection and timely intervention of HAdV infections are critical in clinical practice. The nonspecific clinical manifestations of AdV infections and the challenge of early detection in severe cases necessitate a comprehensive assessment (30). Combining viral load, genotype, and relevant laboratory markers could improve the recognition of severe AdV pneumonitis and treatment outcome (31). Our meta-analysis investigated the association among HAdV load, its genotypic diversity, and the severity of respiratory disease in children. The results indicated that disease severity in children increases with rising viral load, in accordance with other clinical observations and research. In the pediatric population, HAdV infection can manifest as a spectrum of conditions ranging from mild upper respiratory tract infections to severe pneumonia, with the latter associated with higher mortality rates and sequelae. Analyses of the diagnostic efficacy of the B3 and B7 virus subtypes for severe pneumonia in the present study revealed high combined sensitivity and specificity. This suggests a potential role for these subtypes in respiratory infections in children, particularly the development of severe pneumonia. This finding offers a crucial diagnostic reference for clinicians treating children with suspected adenoviral infections, aiding in earlier identification and intervention in severe cases. Furthermore, the real-time qPCR method demonstrated high sensitivity and specificity in detecting these two subtypes, potentially making it more suitable for clinical application. This study provides valuable insights into the role of AdV in childhood respiratory infections and may guide clinical diagnosis and the development of therapeutic strategies. Our findings support integrating qPCR-based HAdV subtyping (particularly for B7) into clinical workflows for children with severe respiratory symptoms, as high viral load and B7 detection correlate with worse outcomes. This enables early risk stratification, guiding decisions on antiviral therapy or intensive care allocation, while minimizing unnecessary interventions in low-risk cases through high specificity.
The main similarities and differences between the present study and other studies are detailed below. Our results suggest that high viral loads are associated with the severity of respiratory illness in children. This finding aligns with the studies by Franz et al. (32) and Martin et al. (33). Moreover, Gao et al. (34) found that Mycoplasma pneumoniae pneumonia was more severe in children with HAdV coinfections than in those with single M. pneumoniae alone. Additionally, Isik et al. (35). reported that a higher initial viral load in adenoviral keratoconjunctivitis may predict inflammatory sequelae. The I2 value of 90% indicates high heterogeneity in the literature, which may be attributed to differences in sample sizes, regions, study designs, and assay methods across studies. Regarding the specificity and sensitivity of B3 and B7 isoforms, our analyses showed high combined sensitivity and specificity, consistent with the study by Wang et al. (36), which found that HAdV-3 and HAdV-7 were the predominant types in children hospitalized with acute respiratory infections in Beijing. The authors showed that HAdV-7 infections caused a more pronounced inflammatory response, severe pulmonary symptoms, and extended hospitalization than HAdV-3, suggesting a need for further clinical typing of HAdV. Although these results are in line with our findings overall, we could not include the data from the study by Wang et al. (36) due to a lack of specific genotyping information for severe pneumonia cases. Li et al. (37) found that 13.8% (76/552) of patients with acute respiratory infections were HAdV-positive, with the prevalence varying by region: 20.1% in north central areas and 8.2% in eastern areas. Xu et al. (38). reported that among AdV-infected children, only 4% had HAdV-55, while 53% had HAdV-7. HAdV-55 infections were mainly detected in March and April, whereas HAdV-7 infections occurred year-round. Our study also found that real-time qPCR showed high sensitivity and specificity in virus detection. These results are in accordance with the studies by Heim et al. (39), Lin et al. (40), and Wong et al. (41), which highlighted the advantages of real-time fluorescence-based qPCR in detecting multiple pathogens. Qiu et al. (42) demonstrated rapid, differential detection and quantitative determination of HAdV serotypes 2, 3, and 7 using triple real-time qPCR, consistent with our findings. We assessed the publication bias using Deeks’ method, finding no significant bias, which lends credibility to our results. This underscores the importance of considering potential bias in epidemiological studies. Our results align with the existing literature, confirming the association among HAdV load, the B3 and B7 subtypes, and the severity of respiratory disease in children. The findings support the importance of real-time qPCR in clinical applications, contributing to a better understanding and management of adenoviral infections in children.
This study had several methodological limitations that may affect the accuracy and reliability of its findings and conclusions. First, the quantity and quality of available literature restrict the value of the findings. Using an observational study, we could not infer causality or completely exclude the influence of confounding factors. Second, study definitions and adjudication criteria varied widely; for example, the definition of a vomiting episode differed among studies, potentially introducing categorical bias, and was not confirmed by subgroup analyses. Additionally, follow-up lengths varied and were sometimes short, with neonatal adverse events possibly being omitting, thus reducing result accuracy. Third, potential publication bias might have affected the generalizability of the results, despite efforts to ensure comprehensive analyses. Publication bias risk also suggests that the effect sizes might have been overestimated, but this was not quantitatively assessed. Finally, the high study heterogeneity was not thoroughly analyzed or corrected for by a random effects model, which could have reduced precision; meanwhile, unrecorded factors such as pregnancy complications might have confounded the results. Considering the multiple subtypes of HAdV, more comprehensiveness analyses beyond only two subtypes could be performed to explore the relationship between a greater array of subtypes and disease severity. We did not adequately account for other factors that may influence disease severity, such as host immune status, comorbidities, and environmental factors. The study’s limitations—including design and definition issues, incomplete and poor-quality data, and unrecorded or underanalyzed confounders—may compromise its accuracy and reliability. Future studies should address these shortcomings, employ more rigorous methodologies, and collect comprehensive, high-quality data to produce more accurate, objective findings to more reliably inform clinical prevention and treatment rationale.
Conclusions
Using meta-analysis, this study thoroughly investigated the association among HAdV load, the B3 and B7 subtypes, and the severity of respiratory disease in children, providing new insights and scientific evidence for clinical treatment. The results showed a significant association between high viral load and increased risk of severe pneumonia, further confirming the importance of viral load in the assessment of disease severity. Meanwhile, this study also found high combined sensitivity and specificity for the B3 and B7 subtypes, suggesting the potential role of these two subtypes in childhood respiratory infections, particularly in the progression to severe pneumonia. In addition, the real-time qPCR method examined in this study showed high sensitivity and specificity for detecting the B3 and B7 virus subtypes, which may be a critical tool for the clinical diagnosis and the development of therapeutic strategies. The study’s scientific value is that it provides a novel perspective in assessing the impact of viral load and specific subtypes on children’s health using quantitative methods, which enriches the existing body of medical knowledge and provides a new direction for future research.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the MOOSE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-2024-627/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-2024-627/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-2024-627/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Costa LF, Da Silveira HL, Queiróz DAO, et al. Respiratory virus infections in hospitalized and non-hospitalized children: determinants of severe course of the disease. J Infect Dev Ctries 2022;16:196-205. [Crossref] [PubMed]
- Tripathi S, Al-Sayyed B, Gladfelter TR. Comparative epidemiology, hospital course, and outcomes of viral respiratory infections in hospitalized pediatric patients. Indian J Med Microbiol 2021;39:24-9. [Crossref] [PubMed]
- Santos JAP, Juanico E, Abello JJ, et al. Surveillance of human adenoviruses in water environments: Assessing the suitability of a locally developed quenching of unincorporated amplification signal reporters-loop-mediated isothermal amplification assay. J Virol Methods 2024;330:115041. [Crossref] [PubMed]
- Nair H, Brooks WA, Katz M, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet 2011;378:1917-30. [Crossref] [PubMed]
- Wang X, Li Y, Shi T, et al. Global disease burden of and risk factors for acute lower respiratory infections caused by respiratory syncytial virus in preterm infants and young children in 2019: a systematic review and meta-analysis of aggregated and individual participant data. Lancet 2024;403:1241-53. [Crossref] [PubMed]
- Videla C, Carballal G, Misirlian A, et al. Acute lower respiratory infections due to respiratory syncytial virus and adenovirus among hospitalized children from Argentina. Clin Diagn Virol 1998;10:17-23. [Crossref] [PubMed]
- Wang H, Zheng Y, Deng J, et al. Molecular epidemiology of respiratory adenovirus detection in hospitalized children in Shenzhen, China. Int J Clin Exp Med 2015;8:15011-7. [PubMed]
- Zhang J, Zhu Y, Zhou Y, et al. Pediatric adenovirus pneumonia: clinical practice and current treatment. Front Med (Lausanne) 2023;10:1207568. [Crossref] [PubMed]
- Resa C, Magro S, Marechal P, et al. Development of an efficient qRT-PCR assay for quality control and cellular quantification of respiratory samples. J Clin Virol 2014;60:270-5. [Crossref] [PubMed]
- Zhong L, Lin J, Dai J. Risk factors for the development of bronchiolitis obliterans in children with severe adenovirus pneumonia: A retrospective study with dose-response analysis. J Med Virol 2020;92:3093-9. [Crossref] [PubMed]
- Abdullah O, Fall A, Klein E, et al. Increased circulation of human adenovirus in 2023: an investigation of the circulating genotypes, upper respiratory viral loads, and hospital admissions in a large academic medical center. J Clin Microbiol 2024;62:e0123723. [Crossref] [PubMed]
- Kim JS, Lee SK, Ko DH, et al. Associations of Adenovirus Genotypes in Korean Acute Gastroenteritis Patients with Respiratory Symptoms and Intussusception. Biomed Res Int 2017;2017:1602054. [Crossref] [PubMed]
- Zeng SZ, Xie LY, Yu T, et al. Persistent viral shedding of human adenovirus type 7 in children with severe pneumonia. J Med Virol 2021;93:4846-55. [Crossref] [PubMed]
- Huanɡ CY, Xianɡ XY, Zhanɡ SY, et al. Monitoring results of human adenovirus and influenza virus in 1,447 children cases of influenza-like illness. Pract Prev Med 2024;31:257-61.
- Sonɡ Z, Wanɡ X, Zhenɡ JL, et al. Study on differentially expressed genes and Chinese medicine prediction of adenovirus infection in children. China Journal of Traditional Chinese Medicine and Pharmacy 2022;37:5425-9.
- Li MH, Liu Q, Zhanɡ Y, et al. Human adenovirus pulmonary infection in 84 children and DNA genotypes. Chinese Journal of Nosocomiology 2022;32:3763-6.
- Siminovich M, Murtagh P. Acute lower respiratory tract infections by adenovirus in children: histopathologic findings in 18 fatal cases. Pediatr Dev Pathol 2011;14:214-7. [Crossref] [PubMed]
- Barrero PR, Valinotto LE, Tittarelli E, et al. Molecular typing of adenoviruses in pediatric respiratory infections in Buenos Aires, Argentina (1999-2010). J Clin Virol 2012;53:145-50. [Crossref] [PubMed]
- Wei X, Guo MY, Jie L, et al. Influencing factors for respiratory tract adenovirus infection in children and subtypes of adenovirus. Chinese Journal of Nosocomiology 2023;33:618-22.
- Esposito S, Zampiero A, Bianchini S, et al. Epidemiology and Clinical Characteristics of Respiratory Infections Due to Adenovirus in Children Living in Milan, Italy, during 2013 and 2014. PLoS One 2016;11:e0152375. [Crossref] [PubMed]
- Fu Y, Tang Z, Ye Z, et al. Human adenovirus type 7 infection causes a more severe disease than type 3. BMC Infect Dis 2019;19:36. [Crossref] [PubMed]
- Goikhman Y, Drori Y, Friedman N, et al. Adenovirus load correlates with respiratory disease severity among hospitalized pediatric patients. Int J Infect Dis 2020;97:145-50. [Crossref] [PubMed]
- Lin MR, Yang SL, Gong YN, et al. Clinical and molecular features of adenovirus type 2, 3, and 7 infections in children in an outbreak in Taiwan, 2011. Clin Microbiol Infect 2017;23:110-6. [Crossref] [PubMed]
- Nguyen DD, Phung LT, Thanh Tran HT, et al. Molecular subtypes of Adenovirus-associated acute respiratory infection outbreak in children in Northern Vietnam and risk factors of more severe cases. PLoS Negl Trop Dis 2023;17:e0011311. [Crossref] [PubMed]
- Papan C, Argentiero A, Adams O, et al. Association of viral load with TRAIL, IP-10, CRP biomarker signature and disease severity in children with respiratory tract infection or fever without source: A prospective, multicentre cohort study. J Med Virol 2023;95:e28113. [Crossref] [PubMed]
- Wanɡ JJ, Duan YL, Ai JH, et al. Epidemiological characteristics of acute lower respiratory tract infection with human adenovirus among children in China from 2017 to 2020. China Preventive Medicine 2024;25:283-8.
- Wo Y, Lu QB, Huang DD, et al. Epidemical features of HAdV-3 and HAdV-7 in pediatric pneumonia in Chongqing, China. Arch Virol 2015;160:633-8. [Crossref] [PubMed]
- Xie LY, Zeng SZ, Yu T, et al. Viral loads in nasopharyngeal aspirates and tracheal aspirates among children hospitalized with invasive ventilation for human adenovirus pneumonia. Virol J 2021;18:238. [Crossref] [PubMed]
- Zhang R, Wang H, Tian S, et al. Adenovirus viremia may predict adenovirus pneumonia severity in immunocompetent children. BMC Infect Dis 2021;21:213. [Crossref] [PubMed]
- Lu L, Zhong H, Su L, et al. Detection and Molecular Characterization of Human Adenovirus Infections among Hospitalized Children with Acute Diarrhea in Shanghai, China, 2006-2011. Can J Infect Dis Med Microbiol 2017;2017:9304830. [Crossref] [PubMed]
- Cao Y, Yang J, Li N, et al. Detection and complete genome sequence analysis of human adenovirus in children with acute diarrhea in Yunnan, China, 2015-2021. Arch Virol 2024;169:34. [Crossref] [PubMed]
- Franz A, Adams O, Willems R, et al. Correlation of viral load of respiratory pathogens and co-infections with disease severity in children hospitalized for lower respiratory tract infection. J Clin Virol 2010;48:239-45. [Crossref] [PubMed]
- Martin ET, Kuypers J, Wald A, et al. Multiple versus single virus respiratory infections: viral load and clinical disease severity in hospitalized children. Influenza Other Respir Viruses 2012;6:71-7. [Crossref] [PubMed]
- Gao J, Xu L, Xu B, et al. Human adenovirus Coinfection aggravates the severity of Mycoplasma pneumoniae pneumonia in children. BMC Infect Dis 2020;20:420. [Crossref] [PubMed]
- Isik P, Harbiyeli II, Ozturk G, et al. The Relationship between Clinical Findings and Viral Load in Adenoviral Keratoconjunctivitis. Jpn J Infect Dis 2022;75:592-6. [Crossref] [PubMed]
- Wang FM, Yang CY, Qian Y, et al. Clinical characteristics of human adenovirus infection in hospitalized children with acute respiratory infection in Beijing. Zhonghua Er Ke Za Zhi 2022;60:30-5. [PubMed]
- Li Y, Zhou W, Zhao Y, et al. Molecular typing and epidemiology profiles of human adenovirus infection among paediatric patients with severe acute respiratory infection in China. PLoS One 2015;10:e0123234. [Crossref] [PubMed]
- Xu L, Liu J, Liu C, et al. Case-control study of the epidemiological and clinical features of human adenovirus 55 and human adenovirus 7 infection in children with acute lower respiratory tract infections in Beijing, China, 2008-2013. BMC Infect Dis 2018;18:634. [Crossref] [PubMed]
- Heim A, Ebnet C, Harste G, et al. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 2003;70:228-39. [Crossref] [PubMed]
- Lin CY, Hwang D, Chiu NC, et al. Increased Detection of Viruses in Children with Respiratory Tract Infection Using PCR. Int J Environ Res Public Health 2020;17:564. [Crossref] [PubMed]
- Wong SSY, Yip CCY, Sridhar S, et al. Comparative evaluation of a laboratory-developed real-time PCR assay and RealStar® Adenovirus PCR Kit for quantitative detection of human adenovirus. Virol J 2018;15:149. [Crossref] [PubMed]
- Qiu FZ, Shen XX, Zhao MC, et al. A triplex quantitative real-time PCR assay for differential detection of human adenovirus serotypes 2, 3 and 7. Virol J 2018;15:81. [Crossref] [PubMed]
(English Language Editor: J. Gray)



