
Clinical features and genetic analysis of epilepsy caused by CLCN4 gene mutation: a case report and literature review
Highlight box
Key findings
• Report a novel chloride voltage-gated channel 4 (CLCN4)-related epilepsy phenotype: a 2-year and 4-month-old male with Gly342Arg variant showed fever-triggered cluster focal seizures, responsive to valproate and lamotrigine, but with mild intellectual disability (ID).
• Literature review of 60 cases: most patients exhibit early-onset seizures (focal/tonic-clonic) and moderate-severe ID. Missense variants appear to correlate with severe phenotypes.
What is known and what is new?
• CLCN4 mutations cause X-linked neurodevelopmental disorders with epilepsy, typically severe.
• The Gly342Arg variant presents fever-sensitive seizures and mild delay, suggesting phenotypic heterogeneity.
What is the implication, and what should change now?
• Therapeutic recommendation: valproate, levetiracetam, and lamotrigine are considered viable therapeutic options.
• Research: investigate CLCN4 variant mechanisms (e.g., Cl− channel dysfunction) for precision medicine.
• Practice: long-term follow-up needed to assess refractory cases.
Introduction
Epilepsy is a chronic brain disorder that affects more than 50 million people worldwide, with an additional 5 million new cases diagnosed each year. It is characterized by abnormal discharges from nerve cells, resulting in recurrent epileptic seizures or abnormal sensations and behaviors, with or without loss of consciousness (1). Various pathological processes in the brain contribute to the onset and development of epilepsy, including defects in ion channels or synapses, structural abnormalities caused by tumors or injuries, infections, autoimmune-inflammatory processes, and metabolic disorders (2). Up to 80% of individuals with epilepsy have an underlying genetic cause, and 1,440 epilepsy-related genes have been identified to date, including those associated with voltage-gated chloride channels (CLCs) (3).
Chloride, the most abundant anion in the body, can induce epileptic seizures by altering the voltage of cell membranes through transmembrane transport, modulating neuronal excitability, and increasing susceptibility to epilepsy (4,5). The chloride voltage-gated channel 4 (CLCN4) gene encodes a voltage-dependent chloride/proton exchanger, which is a 2Cl−/H+ exchanger primarily located in intracellular vesicles (6). Its core function is to maintain endosomal ion homeostasis and vesicle transport (7,8). CLCN4 gene mutations may impair chloride/proton exchange function, disrupting endosomal acidification and ion homeostasis (9-11). These disruptions affect the transport and localization of dendritic ion channels (such as Na+ or K+ channels), causing dendritic excitability dysregulation and morphological abnormalities, which lead to neuronal electrical activity disorders. Clinically, these disturbances often manifest as epilepsy and neurodevelopmental disorders (8,12). Disorders associated with CLCN4 gene variants are clinically rare, with only about 150 cases reported worldwide as of July 2024, and fewer than half of the cases of CLCN4-associated epilepsy have been documented (6-8,10,12-20).
In this study, we retrospectively analyzed the clinical data of a patient with epilepsy caused by a CLCN4 gene variant and reviewed reported cases to summarize the clinical features, genetic characteristics, and treatment outcomes, thereby enhancing clinician awareness of this condition. We present this article in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-2025-157/rc).
Case presentation
A 2-year and 4-month-old male was admitted to the Department of Pediatric Neurology at Chengdu Women’s and Children’s Centre Hospital on March 24, 2021, due to recurrent convulsions. His first episode occurred at 1 year and 11 months of age, characterized by twitching of the mouth accompanied by salivation, loss of consciousness with or without gaze deviation, and limb tonicity, lasting approximately 1 minute and resolving without residual neurological signs. Seizures varied in frequency, occurring from once every few days to five or six times daily, and were easily triggered by fever, often presenting in clusters.
The child was delivered spontaneously with a birth weight of 3,000 g. There was no history of perinatal hypoxic asphyxia, and he achieved normal developmental milestones before the seizure onset. The child was able to lift the head at 2 months, turn over at 4 months, sit alone at 6 months, crawl at 8 months, stand alone at 10 months, walk alone at 12 months, and speak short sentences as well as run and jump at 2 years old. His parents are non-consanguineous and in good health, with an unremarkable family history.
Physical examination revealed a weight of 12 kg, no special facial features, absence of skin depigmentation or café au lait spots, normal muscle strength and tone in the limbs, and negative signs of meningeal irritation and other pathological findings.
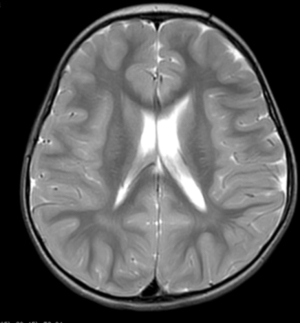
Laboratory findings revealed no abnormalities in blood, urine, or feces, as well as in liver and kidney function, electrolytes, blood glucose, blood ammonia, and blood lactate levels. The electrocardiogram indicated mild T-wave changes. The video electroencephalogram (VEEG) showed a background rhythm of 7–8 Hz. During the interictal period, small spike waves could be observed in the bilateral central regions and central midline regions in the bipolar montage during sleep (Figure 1). No ictal patterns were recorded. Cranial magnetic resonance imaging (MRI) indicated that the left ventricle was slightly wider than its contralateral counterpart (Figure 2).

The patient was diagnosed with epilepsy characterized by focal seizures with impairment of consciousness and was initially treated with oxcarbazepine at a maximum dose of 43 mg/(kg·day), but seizures remained uncontrolled. Subsequently, lamotrigine and sodium valproate were introduced, with gradual dose escalation; oxcarbazepine was subsequently reduced and discontinued. Seizures were progressively controlled, and the patient has been seizure-free for 1 year, currently receiving 24 mg/(kg·day) of sodium valproate and 4.4 mg/(kg·day) of lamotrigine. At present, this child is 5 years and 10 months old, weighing 17 kg (10th to 25th percentile) and measuring 114 cm in height (25th to 50th percentile). He is active, exhibits mild intellectual disability (ID), and demonstrates slightly poorer logical thinking and verbal expression.
Molecular studies
Genomic studies were performed on the peripheral blood of the proband and parents to determine the cause. Genomic DNA was hybridized with the MyGenostics GenCap® Epilepsy Panel V3.0 (603 genes, 2.24 Mb). Libraries were sequenced on Illumina NovaSeq 6000, achieving a mean depth of 621.19× with 99.68% (10×) and 99.43% (20×) target coverage. Variants filtered for rarity [minor allele frequency (MAF) <0.02 in 1000 Genomes/ExAC/EVS] and predicted for pathogenicity using sorting intolerant from tolerant (SIFT), PolyPhen-2, MutationTaster, genomic evolutionary rate profiling (GERP++), and splicing prediction of intronic and exonic variants (SPIDEX) (for splice variants). Pathogenic variants were evaluated following the American College of Medical Genetics and Genomics (ACMG) guidelines (21). Identified pathogenic variant sites were verified by Sanger sequencing.
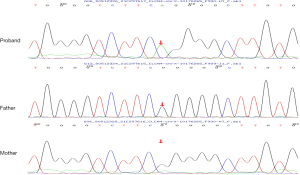
The gene panel analysis revealed that the child had a hemizygous mutation (NM_001830) in the CLCN4 gene, specifically at exon 9, c.1024G>A, which alters amino acid 342 from glycine to arginine. The parents did not carry this variant, suggesting it may be a de novo variant (Figure 3) [pathogenic strong 2 (PS2)_Moderate]. This variant is absent in ExAC, gnomAD, and the Thousand Genomes Asian Population Database, indicating it is a low-frequency variant [pathogenic moderate 2 (PM2)_Supporting]. Moreover, literature reports indicate that the p. Gly342Arg variant in CLCN4’s helix J disrupts ion channel gating by destabilizing the conserved transmembrane domain (steric clashes with Phe339/Leu345), reducing protein expression and impairing heterodimerization (12) [pathogenic strong 3 (PS3)]. The p. Gly342Arg variant (NM_001830.4: c.1024G>A) is classified in ClinVar as uncertain significance (VUS) for X-linked ID (VCV001201131.3), and comprehensive predictions from bioinformatics protein function software, including REVEL, suggested deleterious outcomes, while SIFT, PolyPhen_2, Mutation Taster, and GERP+ all predicted deleterious effects, which further supports disease relevance [pathogenic supporting 3 (PP3)]. According to the 2015 ACMG guidelines, c.1024G>A was classified as a pathogenic variant (PS2_Moderate + PS3 + PM2_Supporting + PP3).
No other pathogenic variants in known epilepsy genes were detected. The SIK1 VUS (maternally inherited) was excluded lacking causality evidence for Developmental and Epileptic Encephalopathy type 30, given the patient’s mild phenotype (controlled epilepsy, no severe delay) and mother’s normal status, with no reports of incomplete penetrance.
Literature review and summary of 60 cases with epilepsy caused by CLCN4
To review the literature on the clinical features of CLCN4-related epilepsy, a comprehensive search was conducted in PubMed, Embase, and Web of Science, up to 7 July 2024, using the following terms: “CLCN4” or “chloride channel 4 protein, human” and “epilepsy” or “Seizure”. Two authors independently screened the literature, excluding duplicate publications by reading the titles and abstracts, and excluding those with incomplete information by reading the full text. A total of 12 articles in English were included (6-8,10,12-14,16-19,22), reporting 59 cases of epilepsy associated with variants in the CLCN4 gene. Including our case, this brings the total to 60 cases, comprising 51 males and 9 females, with 14 patients of Chinese descent. The clinical characteristics of the 60 patients were presented as follows (see Table S1 for details): the majority of patients exhibited neurological and developmental issues, including ID (59/60, 98.3%), language delay (LD) (56/60, 93.3%), and abnormal appearance (31/60, 51.7%). Other notable features included infantile hypotonia (13/60, 21.7%), gastrointestinal symptoms (13/60, 21.7%), and a family history of neurodevelopmental disorders or epilepsy (13/60, 21.7%).
The epilepsy onset age ranged from neonatal period to 16 years, with 48 patients (80.0%) experiencing onset before the age of 3 years. The most common seizure types were focal seizures (32/57, 56.1%) and generalized tonic-clonic (GTC) seizures (22/57, 38.6%). followed by myoclonic (11/57, 19.3%), absence (10/57, 17.5%), spasms (9/57, 15.8%), and tonic seizures (8/57, 14.0%). Atonic, atypical absence, eyelid myoclonus, and myoclonic-atonic seizures were relatively rare. Additionally, nineteen patients (33.3%) were classified into epilepsy syndromes, including infantile spasms (9/19, 47.4%), unclassifiable epileptic encephalopathy (8/19, 42.1%), Lennox-Gastaut syndrome (1/19, 5.3%), and epilepsy of infancy with migrating focal seizures (1/19, 5.3%). Seizures can be triggered by fever in a minority of patients, with two or more seizure types present in 38 cases (67%).
Regarding treatment outcomes, among the 57 cases with medication records, 26 patients (45.6%) were drug-resistant, while 31 patients (54.4%) achieved seizure control with one or more antiseizure medications. The most commonly used effective medications were sodium valproate (10/31, 32.2%), levetiracetam (6/31, 19.4%), and lamotrigine (5/31, 16.1%).
Electroencephalogram (EEG) findings in CLCN4-related epilepsy patients most commonly show focal discharges, with frequent focal spikes or sharp waves, often appearing in the central and central midline regions (13/43, 30.2%); followed by multifocal spikes or sharp waves (11/43, 25.6%); some patients exhibit interictal hypsarrhythmia patterns (6/43, 13.9%), while a few patients show generalized spikes or polyspikes (2/43, 4.7%); some patients present with slowed background activity (5/43, 11.6%).
Cranial MRI findings in these patients include white matter abnormalities: common white matter signal abnormalities, such as delayed myelination and white matter reduction (22/45, 48.9%); ventricular enlargement (14/45, 31.1%); corpus callosum abnormalities: hypoplasia or thinning of the corpus callosum is relatively common (16/45, 35.6%); some patients exhibit cerebral atrophy, manifested as widened sulci and thinned gyri (9/45, 20.0%).
A total of 39 variant sites were involved, with missense variants being the most common (52/60, 86.7%), followed by frameshift variants (4/60, 6.7%), copy number deletions (2/60, 3.3%), and one case each of nonsense variant and intronic splice site variant (1/60, 1.7%). 38 cases were inherited variants (38/60, 63.3%), and 22 cases were de novo variants (22/60, 36.7%). The variant sites with relatively high frequencies were c.1606G>A (8/60, 13.3%), c.823G>A (5/60, 8.3%), c.2152C>T (4/60, 6.7%), and c.1630G>C/A (3/60, 5.0%).
Among the 52 cases with missense variants, 38 presented (73.1%) with moderate to profound developmental disorders, 30 patients (57.7%) had epilepsy of focal origin, 20 (38.5%) were classified as epilepsy syndromes, 25 (48.1%) were drug-resistant, and 5 (9.6%) experienced epilepsy-related deaths. All 4 cases with frameshift variants had mild to moderate ID, with generalized onset seizures that were well-controlled by medication. Of the 2 cases with copy number deletions, 1 had severe ID with multiple seizure types and drug-resistant epilepsy, while the other had mild ID with GTC seizures controlled by monotherapy. The patient with a nonsense variant had severe developmental disorder, focal epilepsy, and was controlled by medication. The patient with an intronic splice site variant had borderline intellectual functioning, unspecified seizure type, and was controlled by medication.
All procedures performed in this study were in accordance with the Declaration of Helsinki and its subsequent amendments. This study was approved by the Ethics Committee of Chengdu Women’s and Children’s Central Hospital (No. 2024-112). Written informed consent was obtained from the patient’s legal guardian for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
Association between CLCN4 gene variants and epilepsy
The CLCN4 gene is located on chromosome Xp22.2 and comprises 13 exons that encode a protein of 760 amino acids. chloride channels and transporters (ClC) proteins form homodimers, each containing distinct ionic pathways within separate subunits, which consist of helical transmembrane structural domains, helical intramembrane structural domains, and cytoplasmic cystathionine beta-synthase (CBS) structural domains. The transmembrane structural domains create the ionic pathways, while the CBS structural domains play a crucial role in influencing the gating and localization of transmembrane proteins, as well as regulating transmembrane components (23,24).
The CLCN4 protein is predominantly expressed during the fetal stage, with significant expression observed in the cranial brain, particularly in the vertebral cells and the Purkinje cell layer of the dentate gyrus and cerebellum. This expression pattern may be associated with ion homeostasis, intracellular vesicular transport, and neuronal differentiation in vivo (4,7). Literature reports indicate a marked reduction in the number and length of dendritic arbors in hippocampal neurons from CLCN4-deficient and CLCN4 knockout mice (13). In contrast, variants of the human CLCN4 gene can result in a rare X-linked neurodevelopmental disorder characterized by varying degrees of ID, psychiatric behavioral abnormalities, and seizures, commonly referred to as Raynaud-Claes syndrome (6,10).
The patient from our study presented with recurrent seizures in early childhood alongside developmental delay, with the CLCN4 gene variant c.1024G>A affecting residue J of the helical transmembrane structural domain and leading to a reduction in CLCN4 protein expression (12). Consequently, this CLCN4 gene variant was identified as the causative factor for this patient’s condition.
Phenotype associated with CLCN4-associated epilepsy
Ninety-eight percent of the patients with CLCN4-associated epilepsy exhibit varying degrees of developmental disorders, primarily moderate to severe developmental delays, with language function impairment being particularly prominent, manifested as language developmental delay or severely impaired expressive ability, such as being able to speak only a few words (8,15,20). This suggests that CLCN4 gene variants have a significant impact on nervous system development, possibly related to neuronal development, migration, or synapse formation (8,12). Literature reports that, compared to CLCN4 gene variant patients without epileptic seizures, patients with epilepsy have more severe ID, and seizure frequency positively correlates with the degree of cognitive decline. Most patients showed no improvement in cognitive function after seizure control, suggesting that CLCN4 dysfunction may directly cause irreversible neurodevelopmental damage (8,12). Our patient’s development did not return to normal levels even after seizure control, further supporting this observation.
Sixty percent of the patients had comorbid emotional and behavioral problems, such as autism spectrum disorder, attention deficit hyperactivity disorder, etc., suggesting that CLCN4 gene variants may affect multiple aspects of the nervous system, leading to abnormalities in cognition, emotion, and behavior (8,20). The presence of comorbidities may have a significant impact on patient prognosis and quality of life, which requires attention in clinical management.
Our literature review indicated that seizure manifestations are diverse, with focal onset and GTC seizures being the most common. Some patients are classified into epilepsy syndromes, indicating that CLCN4 gene variants can lead to multiple types of epileptic seizures (8), and the seizure mechanisms may involve abnormal discharges in various neuronal networks.
Notably, our patient’s cluster seizures with febrile sensitivity differ from the typical focal/GTC seizures predominance in the literature (Table S1), while his preserved motor skills contrast with the frequent gait abnormalities reported in (22). This expands the phenotypic spectrum of CLCN4-related disorders.
The relationship between genotype and phenotype
The CLCN4 gene variant-related epilepsy is most commonly associated with missense variants, followed by frameshift variants. Notably, the variant c.1606G>A/p. Pro369Leu was identified in eight patients from the same family lineage, which has been hypothesized to represent a possible hotspot variant (8,10,13). A review of the reported cases revealed phenotypic differences among patients sharing the same genotype within the same family. Specifically, patients with missense variants exhibited a more severe phenotype compared to those with frameshift variants (8). The former group more frequently presented with moderate-to-very severe developmental delays, primarily of focal seizure onset, along with a higher incidence of epilepsy syndromes, drug-refractory epilepsy, and epilepsy-related fatalities. A study conducted by He et al. in 2021 examined the clinical phenotype and gene function in 20 patients with CLCN4-related epilepsy. The findings revealed that all patients harbored loss-of-function variants, which led to a reduction in ClC-4 currents. However, the study noted an absence of a clear correlation between the variant locus and the severity of the patients’ phenotype (8). An observational study conducted by Palmer et al. (22) in 2023 involving 55 patients with CLCN4-related neurodevelopmental disorders identified a predominance of loss-of-function variants, particularly in males. In contrast, gain-of-function variants were more frequently observed in females and were typically associated with severe growth disturbances, feeding difficulties, and gastrointestinal functioning issues.
However, the relationship between these variants and the epilepsy phenotype remains unclear. Most missense variants impact residues located in the transmembrane or intramembrane regions (4,10). Pathogenic CLCN4 missense variants disrupt the subcellular distribution, stability, and expression of the ClC-4 protein, as well as its ability to form heterodimers with ClC-3 in heterologous expression systems (11). Consequently, it has been proposed that the abnormal protein function resulting from CLCN4 missense variants may be more detrimental than protein deletion or reduced expression levels (8). Nevertheless, the current case, alongside findings from Palmer (22) and others who have reported mild developmental delays and effective epilepsy control in patients with a CLCN4 missense variant, as well as case reports of normal development with a single, well-controlled seizure episode (19). This suggests that the disease phenotype may also be influenced by factors such as genetic background and environmental conditions. Furthermore, the relationship between patient phenotype, gene function, and variant loci requires further investigation.
Treatment response
Currently, there is no targeted drug therapy for this disorder; instead, seizure control, along with symptomatic and rehabilitative interventions, represents the primary treatment approaches. Sodium valproate, levetiracetam, and lamotrigine show some effectiveness in controlling seizures, but specific therapeutic effects vary among individuals, possibly related to factors such as the type of gene variant, seizure form, and neurodevelopmental status (6,8,10,12,19,22). Nearly half of the patients have drug-resistant epilepsy, indicating that the treatment of CLCN4 gene variant-related epilepsy is relatively challenging, possibly due to the complexity of epileptic seizures and the difficulty in controlling abnormal neuronal discharges (6,8,10,12,22). For drug-resistant patients, exploring new treatment methods, such as ketogenic diet and neuromodulation therapy, has important clinical significance.
Strengths and limitations
Our case expands the clinical spectrum of CLCN4-related epilepsy. The c.1024G>A (p. Gly342Arg) variant, though previously reported (12), was observed here in a patient with a distinct cluster seizure phenotype triggered by fever. Unlike most reported cases with moderate-to-severe ID, our patient exhibited only mild developmental delay despite harboring a missense variant, suggesting potential phenotypic variability even within the same variant type. The excellent response to valproate and lamotrigine combination provides further evidence for targeted therapeutic strategies in CLCN4-related epilepsy.
Unfortunately, the patient’s follow-up period was relatively short, lacking long-term prognostic data, which prevented a comprehensive assessment of long-term treatment responses and quality of life in patients with CLCN4 gene variant-related epilepsy. Further deepen research on the pathogenic mechanisms of CLCN4 gene variants and protein function, including effects on Cl− ion channel function and mechanisms of abnormal neuronal network discharge, to provide theoretical basis for developing new treatment methods; explore individualized treatment plans based on gene variants to improve treatment efficacy and patient quality of life.
Conclusions
In summary, our analysis of 60 cases demonstrates that the majority of patients with CLCN4-related epilepsy exhibit neurodevelopmental disorders of varying degrees. Cases can range from a single seizure type that is controllable with monotherapy for focal or generalized epilepsy, to developmental and epileptic encephalopathy with neonatal onset, diverse seizure types, and resistance to multiple medications. Notably, phenotypes associated with missense variants tend to be more severe than those linked to frameshift variants. Early multidisciplinary intervention improves outcomes despite the lack of targeted therapies.
Acknowledgments
We are deeply grateful to the patient and his family who were part of this study. We also want to acknowledge the medical staff involved in those cases and MyGenostics’ support for genetic sequencing of this case.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-2025-157/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-2025-157/prf
Funding: This work was financially supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-2025-157/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki and its subsequent amendments. This study was approved by the Ethics Committee of Chengdu Women’s and Children’s Central Hospital (No. 2024-112). Written informed consent was obtained from the patient’s legal guardian for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- WHO. Epilepsy: a public health imperative. Summary. 2019. Available online: https://iris.who.int/handle/10665/325440
- Burk KC, Kaneko M, Quindipan C, et al. Diagnostic Yield of Epilepsy-Genes Sequencing and Chromosomal Microarray in Pediatric Epilepsy. Pediatr Neurol 2024;150:50-6. [Crossref] [PubMed]
- Zhang MW, Liang XY, Wang J, et al. Epilepsy-associated genes: an update. Seizure 2024;116:4-13. [Crossref] [PubMed]
- Jentsch TJ, Pusch M. CLC Chloride Channels and Transporters: Structure, Function, Physiology, and Disease. Physiol Rev 2018;98:1493-590. [Crossref] [PubMed]
- Ni MM, Sun JY, Li ZQ, et al. Role of voltage-gated chloride channels in epilepsy: current insights and future directions. Front Pharmacol 2025;16:1560392. [Crossref] [PubMed]
- Veeramah KR, Johnstone L, Karafet TM, et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia 2013;54:1270-81. [Crossref] [PubMed]
- Sager G, Yukselmis U, Güzel O, et al. Experience with the Ketogenic Diet in a Boy with CLCN4 Related Neurodevelopmental Disorder. Balkan J Med Genet 2023;26:77-82. [Crossref] [PubMed]
- He H, Guzman RE, Cao D, et al. The molecular and phenotypic spectrum of CLCN4-related epilepsy. Epilepsia 2021;62:1401-15. [Crossref] [PubMed]
- Weinert S, Gimber N, Deuschel D, et al. Uncoupling endosomal CLC chloride/proton exchange causes severe neurodegeneration. EMBO J 2020;39:e103358. [Crossref] [PubMed]
- Palmer EE, Stuhlmann T, Weinert S, et al. De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol Psychiatry 2018;23:222-30. [Crossref] [PubMed]
- Guzman RE, Sierra-Marquez J, Bungert-Plümke S, et al. Functional Characterization of CLCN4 Variants Associated With X-Linked Intellectual Disability and Epilepsy. Front Mol Neurosci 2022;15:872407. [Crossref] [PubMed]
- Sahly AN, Sierra-Marquez J, Bungert-Plümke S, et al. Genotype-phenotype correlation in CLCN4-related developmental and epileptic encephalopathy. Hum Genet 2024;143:667-81. [Crossref] [PubMed]
- Hu H, Haas SA, Chelly J, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 2016;21:133-48. [Crossref] [PubMed]
- Zhou P, He N, Zhang JW, et al. Novel mutations and phenotypes of epilepsy-associated genes in epileptic encephalopathies. Genes Brain Behav 2018;17:e12456. [Crossref] [PubMed]
- Xu X, Lu F, Zhang L, et al. Novel CLCN4 variant associated with syndromic X-linked intellectual disability in a Chinese girl: a case report. BMC Pediatr 2021;21:384. [Crossref] [PubMed]
- Abdulkareem AA, Zaman Q, Khan H, et al. Whole exome sequencing identified five novel variants in CNTN2, CARS2, ARSA, and CLCN4 leading to epilepsy in consanguineous families. Front Genet 2023;14:1185065. [Crossref] [PubMed]
- Rossi J, Russo M, Gobbi G, et al. Developmental and epileptic encephalopathy in a young Italian woman with a de novo missense variant in the CLCN4 gene: A case report. Brain Dev 2023;45:445-50. [Crossref] [PubMed]
- He H, Li X, Guzman GA, et al. Expanding the genetic and phenotypic relevance of CLCN4 variants in neurodevelopmental condition: 13 new patients. J Neurol 2024;271:4933-48. [Crossref] [PubMed]
- Lin ZJ, Li B, Lin PX, et al. Clinical application of trio-based whole-exome sequencing in idiopathic generalized epilepsy. Seizure 2024;116:24-9. [Crossref] [PubMed]
- Li S, Zhang W, Liang P, et al. Novel variants in the CLCN4 gene associated with syndromic X-linked intellectual disability. Front Neurol 2023;14:1096969. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Palmer EE, Pusch M, Picollo A, et al. Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition. Mol Psychiatry 2023;28:668-97. [Crossref] [PubMed]
- Scheel O, Zdebik AA, Lourdel S, et al. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 2005;436:424-7. [Crossref] [PubMed]
- Poroca DR, Pelis RM, Chappe VM. ClC Channels and Transporters: Structure, Physiological Functions, and Implications in Human Chloride Channelopathies. Front Pharmacol 2017;8:151. [Crossref] [PubMed]


