
A child of congenital muscular dystrophy-dystroglycanopathy with a novel variant in the CRPPA gene: a case report and literature review
Highlight box
Key findings
• This case report describes a novel variant (c.1119+2T>G) in the CRPPA gene of an α-dystroglycanopathy patient.
What is known and what is new?
• Dystroglycanopathy is a group of disorders characterized by genetic and phenotypic heterogeneity, affecting the brain, muscles, and eyes, mainly due to defective glycosylation of α-dystroglycan. Eighteen pathogenic genes have been implicated in α-dystroglycanopathies, with the CRPPA (ISPD) gene ranking second.
• We report a novel variant in the CRPPA gene of α-dystroglycanopathy, thereby expanding the phenotypic spectrum of the disease.
What is the implication, and what should change now?
• The discovery of a novel variant in the CRPPA gene enhances our understanding of α-dystroglycanopathy. Future research should focus on functionally validating the effect of c.1119+2T>G, which could further advance studies on the genotype-phenotype correlation.
Introduction
Congenital muscular dystrophy (CMD)-dystroglycanopathy is a rare disorder caused by defective glycosylation of dystroglycan, affecting the brain, muscle, and eyes, resulting from impaired glycosylation of dystroglycan (1). Dystroglycan has two subunits, α-dystroglycan (α-DG) and β-dystroglycan, with α-dystroglycanopathy being the most common. To date, 18 pathogenic genes have been associated with α-dystroglycanopathies (2), with the CDP-L-ribitol pyrophosphorylase A (CRPPA; also known as ISPD) gene ranking second (3). The CRPPA gene on chromosome 7p21.2 spans 334 kb and comprises 10 exons that encode a 451-amino-acid cytidyltransferase enzyme. This enzyme is pivotal in the biosynthesis of CDP-ribitol, which FKTN and FKRP subsequently employ for the transfer of ribose-phosphate groups to α-DG (4). The biallelic loss of the CRPPA gene indirectly impacts the glycosylation of α-DG and leads to α-dystroglycanopathies (5).
α-dystroglycanopathies exhibit genetic heterogeneity, encompassing a wide range of clinical phenotypes, from the lethal Walker-Warburg syndrome (WWS) to the mild form of limb-girdle muscular dystrophy (LGMD). Individuals in the same family with identical genetic variants may present distinct clinical phenotypes (6). Few cases of CMD with mental retardation (CMD-MR), as an intermediate phenotype, have been reported to date. Although the pathogenic mechanisms of CRPPA have been elucidated, a clear genotype-phenotype correlation has yet to be established and warrants further investigation.
In this study, we describe a novel variant in the CRPPA gene identified in a child. The patient exhibited a clinical phenotype consistent with CMD-MR, enhancing the understanding of the genotype-phenotype correlation in α-dystroglycanopathies. We present this case in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-2025-6/rc).
Case presentation
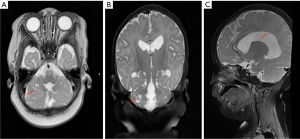
We present a case of a 1-year and 5-month-old girl who exhibited elevated creatine kinase (CK) levels at birth and developed seizures at 1 month of age. Convulsions predominantly presented as repetitive shaking of a single upper or lower limb, occurring 3 to 4 times daily. Physical examination of the child revealed global developmental delay, adequate subcutaneous fat, adducted thumbs, small palpebral fissures, a short interpupillary distance, and limb hypotonia. Laboratory findings showed that serum CK levels typically fluctuated between 2,356 and 9,555 U/L, with significant elevations reaching 54,136 U/L during periods of stress. Interictal video-electroencephalography (VEEG) monitoring revealed sharp waves, multi-spikes, and slow waves predominantly over the left frontal region. During the ictal phase, multifocal discharges were observed in the left anterior region. Brain magnetic resonance imaging (MRI) demonstrated numerous subcortical cysts in the bilateral cerebellar hemispheres and corpus callosum dysplasia (Figure 1). The child was a G2P2, delivered at term by cesarean section due to maternal diabetes during pregnancy, with a birth weight of 4.25 kg. He demonstrates instability in head control and is not yet capable of verbal communication. The parents are non-consanguineous and healthy, with a history of one first-trimester miscarriage. There is no family history of hereditary disorders.
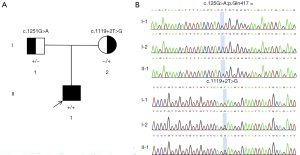
Trio whole-exome sequencing (WES) identified two variants in CRPPA gene [NM_001101426.4:exon9:c.1251G>A (p.Gln417Gln) and NM_001101426.4:intron8:c.1119+2T>G] in the proband, resulting in a compound heterozygous state, with the father and mother being heterozygous carriers of c.1251G>A (p.Gln417Gln) and c.1119+2T>G respectively (Figure 2). The synonymous variant c.1251G>A (p.Gln417Gln) was absent in the gnomAD v4.1.0 database (PM2_Supporting). Multiple splicing prediction algorithms (dbscSNV_ADA_SCORE =0.997, dbscSNV_RF_SCORE =0.920, SpliceAI =0.510) consistently suggest this variant may disrupt the consensus donor splice site in CRPPA intron 9. Reverse transcription polymerase chain reaction (RT-PCR) analysis of messenger RNA (mRNA) confirmed this splicing alteration (PP3) (7). This variant has been reported in the literature in individuals affected with CRPPA-related conditions, five unrelated propands with CMD with confirmed compound heterozygous CRPPA mutations, c.1251G>A/c.789+2T>G, c.1251G>A/c.990delC, c.1251G>A/c.1186G>T, c.1251G>A/c.659A>T, c.1251G>A/exon6-9del (7) (PM3_VeryStrong). According to American College of Medical Genetics and Genomics/Association for Medical Pathology (ACMG/AMP) guidelines, the variant is classified as pathogenic (PM2_Supporting + PM3_VeryStrong + PP3). The c.1119+2T>G was absent in the gnomAD v4.1.0 database (PM2_Supporting) and has not been reported to our knowledge. This variant is located in intron 8, and its adjacent exon 8 has 93 bp, predicted to likely escape NMD (nonsense-mediated mRNA decay) but may result in a truncated protein of <10%. No pathogenic missense variants have been reported within this exon so far (PVS1_Moderate). The variant was detected in compound heterozygosity with the pathogenic variant c.1251G>A (p.Gln417Gln) (PM3). According to ACMG/AMP guidelines, the variant is classified as VUS (PVS1_Moderate + PM2_Supporting + PM3).
The diagnosis of CMD-MR was established based on characteristic clinical features (muscle weakness, hypotonia, normal ocular structure), markedly elevated serum CK levels, neuroimaging findings (cerebellar subcortical microcysts and corpus callosum dysgenesis), electroclinical evidence of focal epilepsy, and the identification of biallelic pathogenic variants in the CRPPA gene. The patient was treated with levetiracetam (25 mg/kg/day) from 1 month of age, achieving seizure-free status. Follow-up VEEG demonstrated a reduction in interictal discharges, with no seizures observed. Levetiracetam was discontinued at 1 year of age without medical supervision, and the patient remained seizure-free. No specific intervention has been provided for the developmental delay, which has not improved.
All procedures performed in this study were in accordance with the ethical standards of Institutional Review Board of The Second Hospital of Shandong University (approval No. KYLL-2022D017) and with the Declaration of Helsinki and its subsequent amendments. Written informed consent was obtained from the patient’s legal guardians for the publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
The pathogenesis of α-dystroglycanopathy is attributed to impaired glycosylation of α-DG. α-DG primarily functions to link the dystrophin-glycoprotein complex to extracellular matrix proteins, thereby stabilizing muscle structure and function (2). Loss of α-DG glycosylation in muscular dystrophies disrupts its linkage to extracellular proteins, thereby contributing to disease pathogenesis. It is currently believed that 18 genes are associated with α-dystroglycanopathy. FKRP (8), POMGNT1 (9), and POMT1 (10) are recognized as the most common pathogenic genes for α-dystroglycanopathy in Europe and Asia. With the identification of new variants, the CRPPA gene has emerged as the second frequent cause.
In this study, we report a case of α-dystroglycanopathy caused by compound heterozygous mutations in the CRPPA gene, one of which is a novel variant. The c.1251G>A is a synonymous variant inherited from her father, which affects splicing. RT-PCR analysis of mRNA has confirmed this splicing alteration (7). This variant has thus far been reported exclusively in the Chinese population and is regarded as a hotspot mutation within this population (1,11). The c.1119+2T>G is a classic splice site variant inherited from her mother, which has not been reported to our knowledge. The ACMG predicted the variant escapes NMD and may produce a truncated protein (<10%). Although the pathogenicity of the c.1119+2T>G remains unvalidated by functional assays, the adjacent c.1120-1G>T variant is pathogenic, confirmed by CRPPA complementation in fibroblasts (12).
α-dystroglycanopathies exhibit considerable genetic heterogeneity. However, the mechanism between genotype and clinical phenotype remains unclear. To date, only nine cases of CMD-MR have been reported (1,9,13-16). Our case involves both muscular and cerebral involvement, with no ocular manifestations. She exhibits significant impairment in both motor and intellectual functions. The clinical phenotype strongly supports a diagnosis of CMD-MR, further contributing to the growing case series of CMD-MR. In addition, we have compiled 72 mutation sites of 63 cases, including the new variation we identified (Table 1). We find cases carrying either a missense, synonymous, or in-frame variant were more frequently associated with milder LGMD phenotypes (21/45), while those lacking such variants predominantly presented with severe WWS (13/18). The pathogenicity of variants is correlated with the extent of protein dysfunction. Mutations in the C-terminal cytidyltransferase domain, especially truncating mutations, are more likely to reduce enzyme activity and result in disease. For example, c.1114_1116del (p.Val372del) causes a mild phenotype due to the loss of a single C-terminal residue, while c.1186G>T (p.Glu396*) leads to a larger C-terminal deletion, resulting in a more severe phenotype (33). In our case, the c.1251G>A and c.1119+2T>G variants are likely to impact the C-terminal cytidyltransferase domain, potentially contributing to CMD-MR. However, further functional validation is needed to confirm these findings.
Table 1
| Group | Variant 1 | Variant 2 | Disease | Article | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Exon | Protein | Origin | Location | Exon | Protein | Origin | ||||
| Group 1 (n=45) | c.5A>T | 1 | Glu2Val | Mother | c.505A>T | 2 | Lys169* | Father | MEB | Song et al. (1) | |
| c.79A>C | 1 | Thr27Pro | Novel | c.1218T>G | 9 | Ile406Met | Novel | LGMD | Wu et al. (17) | ||
| c.157G>A | 1 | Ala53Thr | NA | c.1183A>T | 9 | Arg395* | NA | LGMD | Cirak et al. (18) | ||
| c.161G>T | 1 | Gly54Ala | Mother | c.161G>T | 1 | Gly54Ala | Father | LGMD | Tasca et al. (19) | ||
| c.277-279del | 2 | Ile93del | NA | c.789+2T>G | – | – | NA | WWS | Willer et al. (12) | ||
| c.340C>G | 2 | His114Asp | Mother | c.340C>G | 2 | His114Asp | Father | CMD without MR | Song et al. (1) | ||
| c.346C>T | 2 | Arg116Cys | NA | c.947C>A | 7 | Thr316Lys | NA | LGMD | Sframeli et al. (9) | ||
| c.364G>C | 2 | Ala122Pro | Mother | c.802C>T | 5 | Arg268* | Father | MEB | Roscioli et al. (3) | ||
| c.367G>A | 2 | Gly123Arg | Mother | c.367G>A | 2 | Gly123Arg | Father | LGMD | Baranello et al. (6) | ||
| c.377G>A | 2 | Arg126His | Mother | c.53dup | 1 | Ser19Glufs*97 | Father | WWS | Cirak et al. (18) | ||
| c.377G>A | 2 | Arg126His | NA | c.2T>G | 1 | Met1Arg | NA | LGMD | Sframeli et al. (9) | ||
| c.377G>A | 2 | Arg126His | NA | c.2T>G | 1 | Met1Arg | NA | CMD-MR | Sframeli et al. (9) | ||
| c.446C>T | 2 | Pro149Leu | NA | c.643C>T | 3 | Gln215* | NA | CMD without MR | Cirak et al. (18) | ||
| c.457A>T | 2 | Ile153Phe | NA | – | 6-9del | – | NA | LGMD | Song et al. (1) | ||
| c.458T>C | 2 | Ile153Thr | Novel | c. 535-3C>G | – | – | Novel | CMD without MR | Ceyhan-Birsoy et al. (20) | ||
| c.464A>G | 2 | His155Arg | Father | c.712A>G | 4 | Thr238Ala | Mother | CMD-MR | Song et al. (1) | ||
| c.550C>T | 3 | Arg184Gly | Mother | c.984G>T | 7 | Glu328His | Father | LGMD | Yang et al. (21) | ||
| c.538G>A | 3 | Ala180Thr | NA | c.538G>A | 3 | Ala180Thr | NA | LGMD | Song et al. (1) | ||
| c.605C>T | 3 | Ser202Leu | NA | c.165dup | 1 | Cys56Valfs*60 | NA | LGMD | Johnson et al. (22) | ||
| c.613C>T | 3 | Arg205Cys | Father | c.747C>A | 4 | Cys249* | Mother | CMD-MR | Chen et al. (15) | ||
| c.614G>A | 3 | Arg205His | Mother | – | 9-10del | – | Father | WWS | Czeschik et al. (23) | ||
| c.641C>T | 3 | Pro214Leu | NA | chr7:16312789-16323521del | – | – | NA | WWS | Alharbi et al. (24) | ||
| chr7:16415756T>G | 3 | Gln215His | Father | chr7:16415756T>G | 3 | Gln215His | Mother | CMD-MR | Biswal et al. (13) | ||
| c.647C>A | 3 | Ala216Asp | Mother | c.647C>A | 3 | Ala216Asp | Father | WWS | Roscioli et al. (3) | ||
| c.659A>G | 3 | Asp220Val | Mother | c.1251G>A | 9 | Gln417 = | Father | CMD without MR | Song et al. (1) | ||
| c.676 T>C | 3 | Tyr226His | NA | c.836-5T>G | – | – | NA | LGMD | Magri et al. (25) | ||
| c.677A>G | 3 | Tyr266Cys | NA | c.53dup | 1 | Ser19Glufs*97 | NA | LGMD | Cirak et al. (18) | ||
| c.1104-1106del | 8 | V372del | Novel | c.1270del | 10 | Glu424Argfs*3 | Novel | LGMD | Hu et al. (26) | ||
| c.1106T>G | 8 | Val369Gly | Mother | c.674del | 3 | Ala225Aspfs*21 | Father | WWS | Chen et al. (15) | ||
| c.1114-1116del | 8 | Val373del | NA | c.1183A>T | 9 | Arg395* | NA | LGMD | Cirak et al. (18) | ||
| c.1114-1116del | 8 | Val373del | Mother | c.1114-1116del | 8 | Val373del | Father | LGMD | Tasca et al. (19) | ||
| c.1114-1116del | 8 | Val373del | NA | c.1027-10G>A | – | – | NA | LGMD | Song et al. (1) | ||
| c.1114-1116del | 8 | Val373del | Mother | c.1026+1G>A | – | – | Father | LGMD | Song et al. (1) | ||
| c.1114-1116del | 8 | Val373del | NA | c.1354T>G | 10 | *452Arg | NA | LGMD | Westra et al. (27) | ||
| c.1124A>G | 9 | His375Arg | Mother | c.1026+1G>A | – | – | Father | LGMD | Song et al. (1) | ||
| C.1231C> T | 9 | Leu411Phe | Father | – | 9del | – | Mother | LGMD | Huang et al. (28) | ||
| c.1251G>A | 9 | Gln417 = | Father | c.1119+2T>G | – | – | Mother | CMD-MR | Our report | ||
| c.1251G>A | 9 | Gln417 = | Mother | c.1186G>A | 8 | Glu396* | Father | CMD-MR | Song et al. (1) | ||
| c.1251G>A | 9 | Gln417 = | Mother | c.789+2T>G | – | – | Father | MEB | Song et al. (1) | ||
| c.1251G>A | 9 | Gln417 = | Father | – | 6-9del | – | Mother | CMD-MR | Song et al. (1) | ||
| c.1251G > A | 9 | Gln417 = | Mother | c.789+2T>A | – | – | Father | WWS | Lin et al. (11) | ||
| c.1251G>A | 9 | Gln417 = | Father | c.990del | 7 | IIe331Serfs*2 | Mother | MEB | Song et al. (1) | ||
| c.1354T>A | 10 | *452Arg | NA | c.1354T>A | 10 | *452Arg | NA | WWS | Willer et al. (12) | ||
| c.1354T>A | 10 | *452Arg | NA | c.1315G>T | 10 | Glu439* | NA | CMD-MR | Meng et al. (16) | ||
| c.1354T>G | 10 | *452Arg | NA | c.184del | 1 | Val62Serfs*29 | NA | LGMD | Nallamilli et al. (8) | ||
| Group 2 (n=18) | c.184del | 1 | Val62Serfs*29 | NA | c.790-1G>C | – | – | NA | WWS | Wojcik et al. (29) | |
| c.217G>T | 1 | Glu73* | Novel | c.217G>T | 1 | Glu73* | Novel | WWS | Bayram et al. (30) | ||
| c.258-1G>A | – | – | NA | c.716-719del | 1 | Glu239Valfs*6 | NA | WWS | Sframeli et al. (9) | ||
| c258-1G>C | – | – | Novel | c.505A>T | 2 | Lys169* | Novel | LGMD | Park et al. (31) | ||
| c.550C>T | 3 | Arg184* | NA | c.550C>T | 3 | Arg184* | NA | WWS | Alharbi et al. (24) | ||
| c.627-628del | 3 | Arg209fs*3 | NA | c.627-628del | 3 | Arg209fs*3 | NA | CMD-MR | Marangoni et al. (14) | ||
| c.643C>T | 3 | Gln215* | NA | – | 9-10del | – | NA | WWS | Willer et al. (12) | ||
| c.790-1G>C | – | – | NA | c.790-1G>C | – | – | NA | WWS | Alharbi et al. (24) | ||
| c.832A>T | 5 | Lys278* | Mother | c.832A>T | 5 | Lys278* | Father | MEB | Roscioli et al. (3) | ||
| c.836-?_1119+?dup | 6-8 | Val374Rfs*8 | NA | c.836-?_1119+?dup | 6-8 | Val374Rfs*8 | NA | LGMD | Cirak et al. (18) | ||
| c.1120-1G>T | – | – | NA | c.1120-1G>T | – | – | NA | WWS | Willer et al. (12) | ||
| c.1186G>T | 9 | Glu396* | Mother | c.1186G>T | 9 | Glu396* | Father | WWS | Roscioli et al. (3) | ||
| – | 3del(a) | – | NA | – | 3del(a) | – | NA | WWS | Willer et al. (12) | ||
| – | 3-5del | – | NA | – | 3-5del | – | NA | WWS | Roscioli et al. (3) | ||
| – | 6-8del | – | Mother | – | 6-8del | – | Father | WWS | Roscioli et al. (3) | ||
| – | 6-9del | – | Mother | c.1114-1116del | 8 | Val373del | Father | LGMD | Yang et al. (21) | ||
| – | 9-10del | – | Mother | – | 9-10del | – | Father | WWS | Roscioli et al. (3) | ||
| – | 2-10del | – | NA | – | 2-10del | – | NA | WWS | Trkova et al. (32) | ||
Group 1: 45 cases, each carrying at least one missense, synonymous, or in-frame variant. Group 2: 18 cases, with neither variant being missense, synonymous, or in-frame. *, stop codon; a: g., (16,401,191–16,406,273) (16,409,318–16,431,594)del. CMD, congenital muscular dystrophy; LGMD, limb-girdle muscular dystrophy; MEB, muscle-eye-brain disease; MR, mental retardation; NA, not available; WWS, Walker-Warburg syndrome.
Currently, there is no curative treatment for α-dystroglycanopathies. In our case, oral levetiracetam has been administered to manage epileptic seizures, but no effective therapies are available to address the motor and intellectual developmental impairments. Increasing ribitol levels may represent a potential therapeutic strategy for patients with CRPPA-related α-dystroglycanopathy (4). Further elucidation of CRPPA’s structural dynamics and substrate interaction mechanisms may accelerate targeted therapy development for this subset of α-dystroglycanopathies.
Conclusions
In summary, we identified two variants in the CRPPA gene, including the novel variant c.1119+2T>G. The patient exhibited clinical features consistent with CMD-MR, thereby expanding the phenotypic spectrum associated with α-dystroglycanopathies.
Acknowledgments
We express our gratitude to Shanghai Enyuan Medical Laboratory Co., Ltd. for providing technical assistance.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-2025-6/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-2025-6/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-2025-6/coif). All authors state that Shanghai Enyuan Medical Laboratory Co., Ltd. has provided technical assistance services. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of Institutional Review Board of The Second Hospital of Shandong University (approval No. KYLL-2022D017) and with the Declaration of Helsinki and its subsequent amendments. Written informed consent was obtained from the patient’s legal guardians for the publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Song D, Dai Y, Chen X, et al. Genetic variations and clinical spectrum of dystroglycanopathy in a large cohort of Chinese patients. Clin Genet 2021;99:384-95. [Crossref] [PubMed]
- Liu S, Su T, Xia X, et al. Native DGC structure rationalizes muscular dystrophy-causing mutations. Nature 2025;637:1261-71. [Crossref] [PubMed]
- Roscioli T, Kamsteeg EJ, Buysse K, et al. Mutations in ISPD cause Walker-Warburg syndrome and defective glycosylation of α-dystroglycan. Nat Genet 2012;44:581-5. [Crossref] [PubMed]
- Gerin I, Ury B, Breloy I, et al. ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto α-dystroglycan. Nat Commun 2016;7:11534. [Crossref] [PubMed]
- Yang H, Manya H, Kobayashi K, et al. Analysis of phenotype, enzyme activity and genotype of Chinese patients with POMT1 mutation. J Hum Genet 2016;61:753-9. [Crossref] [PubMed]
- Baranello G, Saredi S, Sansanelli S, et al. A novel homozygous ISPD gene mutation causing phenotype variability in a consanguineous family. Neuromuscul Disord 2015;25:55-9. [Crossref] [PubMed]
- Song D, Fu X, Ge L, et al. A splice site mutation c.1251G>A of ISPD gene is a common cause of congenital muscular dystrophy in Chinese patients. Clin Genet 2020;97:789-90. [Crossref] [PubMed]
- Nallamilli BRR, Chakravorty S, Kesari A, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol 2018;5:1574-87. [Crossref] [PubMed]
- Sframeli M, Sarkozy A, Bertoli M, et al. Congenital muscular dystrophies in the UK population: Clinical and molecular spectrum of a large cohort diagnosed over a 12-year period. Neuromuscul Disord 2017;27:793-803. [Crossref] [PubMed]
- Graziano A, Bianco F, D'Amico A, et al. Prevalence of congenital muscular dystrophy in Italy: a population study. Neurology 2015;84:904-11. [Crossref] [PubMed]
- Lin L, Zhang Y, Pan H, et al. Clinical and genetic characteristics and prenatal diagnosis of patients presented GDD/ID with rare monogenic causes. Orphanet J Rare Dis 2020;15:317. [Crossref] [PubMed]
- Willer T, Lee H, Lommel M, et al. ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nat Genet 2012;44:575-80. [Crossref] [PubMed]
- Biswal S, Panigrahi D, Mohakud NK, et al. A Child of Congenital Muscular Dystrophy-Dystroglycanopathy with Homozygous Missense Variation in Exon 3 of the ISPD Gene: A Rare Case from Odisha. Adv Biomed Res 2020;9:70. [Crossref] [PubMed]
- Marangoni M, Smits G, Ceysens G, et al. Implementation of fetal clinical exome sequencing: Comparing prospective and retrospective cohorts. Genet Med 2022;24:344-63. [Crossref] [PubMed]
- Chen M, Chen J, Wang C, et al. Clinical application of medical exome sequencing for prenatal diagnosis of fetal structural anomalies. Eur J Obstet Gynecol Reprod Biol 2020;251:119-24. [Crossref] [PubMed]
- Meng L, Pammi M, Saronwala A, et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr 2017;171:e173438. [Crossref] [PubMed]
- Wu L, Brady L, Shoffner J, et al. Next-Generation Sequencing to Diagnose Muscular Dystrophy, Rhabdomyolysis, and HyperCKemia. Can J Neurol Sci 2018;45:262-8. [Crossref] [PubMed]
- Cirak S, Foley AR, Herrmann R, et al. ISPD gene mutations are a common cause of congenital and limb-girdle muscular dystrophies. Brain 2013;136:269-81. [Crossref] [PubMed]
- Tasca G, Moro F, Aiello C, et al. Limb-girdle muscular dystrophy with α-dystroglycan deficiency and mutations in the ISPD gene. Neurology 2013;80:963-5. [Crossref] [PubMed]
- Ceyhan-Birsoy O, Talim B, Swanson LC, et al. Whole Exome Sequencing Reveals DYSF, FKTN, and ISPD Mutations in Congenital Muscular Dystrophy Without Brain or Eye Involvement. J Neuromuscul Dis 2015;2:87-92. [Crossref] [PubMed]
- Yang H, Cai F, Liao H, et al. Case Report: ISPD Gene Mutation Leads to Dystroglycanopathies: Genotypic Phenotype Analysis and Treatment Exploration. Front Pediatr 2021;9:710553. [Crossref] [PubMed]
- Johnson K, Bertoli M, Phillips L, et al. Detection of variants in dystroglycanopathy-associated genes through the application of targeted whole-exome sequencing analysis to a large cohort of patients with unexplained limb-girdle muscle weakness. Skelet Muscle 2018;8:23. [Crossref] [PubMed]
- Czeschik JC, Hehr U, Hartmann B, et al. 160 kb deletion in ISPD unmasking a recessive mutation in a patient with Walker-Warburg syndrome. Eur J Med Genet 2013;56:689-94. [Crossref] [PubMed]
- Alharbi S, Alhashem A, Alkuraya F, et al. Neuroimaging manifestations and genetic heterogeneity of Walker-Warburg syndrome in Saudi patients. Brain Dev 2021;43:380-8. [Crossref] [PubMed]
- Magri F, Colombo I, Del Bo R, et al. ISPD mutations account for a small proportion of Italian Limb Girdle Muscular Dystrophy cases. BMC Neurol 2015;15:172. [Crossref] [PubMed]
- Hu X, Li N, Xu Y, et al. Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet Med 2018;20:1045-53. [Crossref] [PubMed]
- Westra D, Schouten MI, Stunnenberg BC, et al. Panel-Based Exome Sequencing for Neuromuscular Disorders as a Diagnostic Service. J Neuromuscul Dis 2019;6:241-58. [Crossref] [PubMed]
- Huang J, Miao WH, Guo XF, et al. Diagnosis and genetic testing analysis of limb-girdle muscular dystrophy type 2U caused by a compound heterozygous mutation in the ISPD gene. Yi Chuan 2023;45:536-42. [Crossref] [PubMed]
- Wojcik MH, Schwartz TS, Thiele KE, et al. Infant mortality: the contribution of genetic disorders. J Perinatol 2019;39:1611-9. [Crossref] [PubMed]
- Bayram N, Bayram AK, Per H, et al. Analysis of genotype-phenotype correlation in Walker-Warburg syndrome with a novel CRPPA mutation in different clinical manifestations. Eur J Ophthalmol 2022;32:NP71-6. [Crossref] [PubMed]
- Park HJ, Jang H, Kim JH, et al. Discovery of pathogenic variants in a large Korean cohort of inherited muscular disorders. Clin Genet 2017;91:403-10. [Crossref] [PubMed]
- Trkova M, Krutilkova V, Smetanova D, et al. ISPD gene homozygous deletion identified by SNP array confirms prenatal manifestation of Walker-Warburg syndrome. Eur J Med Genet 2015;58:372-5. [Crossref] [PubMed]
- Riemersma M, Froese DS, van Tol W, et al. Human ISPD Is a Cytidyltransferase Required for Dystroglycan O-Mannosylation. Chem Biol 2015;22:1643-52. [Crossref] [PubMed]


