Genetics of hereditary neurological disorders in children
Introduction
According to Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), there are five age stages of neurodevelopment (neonatal, infancy, toddler, childhood, adolescence) incorporated in the terminology of paediatrics (1). Neurological disorders occurred at age from preterm neonatal to late adolescence (19-21 years old) are paediatric neurological disorders, which disease spectrum has considerable overlap with adult neurological disorders. Hereditary neurological disorders (HNDs) are relatively common in paediatric neurological practice. HNDs are a group of genetic diseases, most of which with a Mendelian inheritance affecting neurological system. Although HNDs could occur in any age, the initial symptoms of most HNDs are presented in childhood and congregate in a certain age group. Congenital malformation, if present, could be identified during preterm and post neonatal periods. Developmental retardation is often first suspected in infants (1 month to 2 years), when the developmental milestones could not be reached. Clinical intervention is often taken in place during child (2-12 years old) and adolescence (12-21 years old) age for HNDs (1,2).
In most HNDs, there are four means to guide clinical classification: (I) the earliest clinical signs refer to one neuroanatomical region or pathology specific to that disease; (II) the age of onset for the clinical signs and symptoms; (III) the mode of inheritance; (IV) other extra-neural signs and symptoms, such as the presence of specific signs involving the eyes, skin, connective tissues, or visceral organs, etc(3). The ultimate goal of HND diagnosis is gene diagnosis, although neuroimaging, biochemical analyses of body fluids, etc., would also assist with accurate diagnosis. Just as a set of clinical features could be resulted from mutations in different genes, mutations in a single gene can manifest with different clinical phenotypes. The same gene mutations causing different clinical phenotypes indicate the presence of environmental and genetic modifiers, and similar therapeutic approaches shall be employed in the spectrum of HNDs caused by the single gene. This review will discuss clinical and genetic perspectives of the more commonly encountered HNDs.
Genetic investigation is a complex area. Karyotyping has been widely used to detect gross copy number variants for over 50 years. Sanger sequencing has been the key platform for gene discovery and genetic diagnosis for over three decades. However, due to its limited capacity, Sanger sequencing can only sequence genes sequentially and is labour intensive and time consuming. In contrast, next generation sequencing (NGS) has the capacity to sequence multiple genes even full exons of human genome simultaneously and provides a powerful tool in investigation of complex HNDs. The costing for NGS is expected to be significantly reduced, and its routine clinical application for HNDs can be envisioned in the future (4). Through applications of NGS, genotyping chips and comparative genome hybridization array (CGH), chromosomal structural variation or gene copy number variations (CNVs) have been identified for many HNDs (5). As the rapid advances of genomic technologies, novel genes for HNDs have continually been identified (6,7). Hence, paediatricians and especially paediatric neurologists are facing great challenges with regard to appropriate genetic testing referral, understanding genetic reports and genetic counselling. This review will equip clinicians to better handle these challenges.
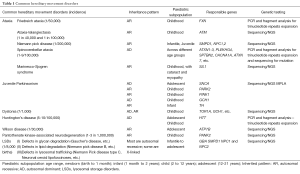
Genetics of paediatric hereditary movement disorders
In paediatric practice, a hereditary movement disorder is often named after the initial symptoms and also a classic adult-onset hereditary movement disorders could present in childhood.
Ataxia is a non-specific clinical manifestation of lack voluntary coordination of muscle movement. Hereditary ataxia could result from neurodegeneration from spine (such as Friedreich ataxia) or cerebellum (such as ataxia-telangiectasia, Niemann Pick disease, Marinesco-Sjögren syndrome), and most cases feature both to some extent (such as spinocerebellar ataxia) (Table 1). Friedreich’s ataxia is caused by expanded GAA repeats in the first intron of FXN gene, which is located on chromosome 9 and encodes the protein frataxin. The mutation causes gene silence and leads to frataxin functional deficiency. Ataxia-telangiectasia (A-T, also referred to as Louis-Bar syndrome) is caused by a defect in the ATM gene, which encodes a cellular sensor protein detecting DNA double-strand breaks and responsible for DNA repair. ATM is a large gene, composed of 66 exons spreading across 150 kb on chromosome 11q. Niemann Pick disease is one of lysosomal storage diseases (LSDs). A total of 36 types of spinocerebellar ataxia have been identified so far, and most of them are caused by CAG trinucleotide repeats expansion in the respective genes, resulting in polyglutamine accumulation (PolyQ disorders) (3,8). Genetic testing guidelines for patients with ataxias are proposed by European Federation of Neurological Societies (9).
Full table
Juvenile Parkinsonism (JP) refers to patients with tremor, bradykinesia and rigidity symptoms before age 21. Hereditary cause is predominant in this age group and Parkinsonism is often accompanied by other features, such as dystonia, seizures and ataxia. Five genes are the major responsible causes for patients primarily present with JP (Table 1) (10). Alpha-synuclein (SNCA), parkin (PARK2) and PINK1 have been recognised to cause Parkinson’s disease, while GCH1 and TH are genes also causing dystonia (seeing Table 1).
Dystonia is a movement disorder characterised by involuntary muscle contractions that cause twisting and repetitive movement or abnormal postures, or both, and maybe tremulous (11). According to the inheritance pattern, hereditary dystonia is classified into four forms: autosomal dominant (AD) [such as torsin family 1 member A (TOR1A); ATPase alpha3 polypeptide (ATP1A3)], autosomal recessive (AR) [such as tyrosine hydroxylase (TH)], X-linked recessive (such as TATA box binding protein associated factor, TAF1), and mitochondria (such as Leigh syndrome) (11). So far, about 20 genes have been identified to cause dystonia. For those children with dopa-responsive dystonia (Segawa’s dystonia), mutation screening for genes, encoding products involved in several neurotransmitter dopamine metabolic pathways, such as TH, sepiapterin reductase, etc., shall be performed.
Huntington disease (HD) is the most common genetic cause of abnormal involuntary writhing movements and cerebellar symptoms. Cognitive decline to severe mental deterioration, speech and language delay, psychiatric problems and rapid decline often occur. Epileptic seizures, unique to the youngest onset group, are present in 30-50% of those with onset of HD before age ten years. In teenagers, symptoms are more similar to adult HD with the characteristic chorea. About 6% HD cases have their symptoms start before age of 21 with akinetic-rigid syndrome (Westphal variant HD). Expanded trinucleotide repeat in Huntingtin gene (HTT) is the genetic cause of HD, which results in polyglutamine accumulation. Individuals carrying 27 to 35 CAG repeats in the HTT gene are at risk to develop HD. As the gene is passed from parent to child, the size of the CAG trinucleotide repeat may increase into the range associated with HD (typical 40 to 50 CAG repeats). The next generation of HD patients may have children with juvenile form of HD with more than 60 CAG repeats, and with childhood onset often exhibiting a repeat size of over 80 (12). The repeats size increases in the next generation, and a larger number of repeats is usually associated with an earlier onset of signs and symptoms, so called genetic anticipation, which is an important feature in HD (13).
Wilson disease is caused by mutations in ATP7B gene, which plays an important role in copper transportation and metabolism. About half patients with Wilson disease will develop neurological and psychiatric symptoms in their 20s. Due to the subtleness of psychiatric problems and neurological symptoms such as parkinsonism, ataxia or dystonia are the prominent complains. Kayser-Fleischer ring (KF ring) due to cooper deposition in cornea is often seen in patients with neurological symptoms. Liver is another main organ affected in Wilson disease, and liver dysfunction is an early sign in children (3).
Pantothenate kinase-associated neurodegeneration (PKAN) is a neurodegenerative disorder with initial symptoms of dystonia or Parkinsonism, and mostly occurs before age 10 due to iron accumulation in the brain. Night blindness is an early sign of PKAN resulting from iron deposition in the retina. Deletion or missense mutations of PANK2 gene lead to deficiency of its protein product, pantothenate kinase 2, results in cell toxicity (3).
LSDs are a group of systematic disorders with brain and other organs affected. Children with LSDs can present with variable symptoms, such as developmental delay, movement disorders, seizures and cognitive impairment, etc. Most LSDs follow AR inheritance pattern, resulting from defective genes encoding specific lysosomal enzymes (14) (Table 1).
Genetics of developmental & neuropsychiatric disorders
Neuropsychiatric disorders include a variety of clinical features, such as developmental delay, intellectual disability, autism spectrum disorders (ASDs) and cognitive dysfunction, etc. Many intellectual developmental disorders, such as attention deficit hyperactivity disorder, are due to complex genetics contribution, complication during pregnancy or during birth, or environmental factors (such as exposure to lead), etc. Many neuropsychiatric disorders follow Mendelian genetic inheritance pattern. This section will only discuss genetic related neuropsychiatric disorders.
Down syndrome (DS), also known as trisomy 21, is the most common chromosome abnormality in human. Affected children have problems with thinking and learning, impulsive behaviour, speech delay and physical disabilities. A particular set of eight physical characteristics could assist with early diagnosis of a child with DS (Table 2) (15). DS is caused by the presence of an extra copy of chromosome 21 and additional dose of a part of chromosome 21, leading to an overexpression of many genes on chromosome 21. Non-invasive prenatal diagnosis of DS from cell-free DNA in maternal circulation has been extensively used now.

Full table
ASDs affect around one in 150 children, and are characterized by varying degrees of limitations in communication and social interaction and atypical, repetitive behaviours with an onset before three years of age. Advances in clinical testing technology and proper clinical referral considerations have increased the diagnostic yield from 6-10% a few years ago to 30-40% (16). To assure an accurate diagnosis of autism before proceeding with genetic investigation is very important to achieve the high genetic diagnostic yield (16). With application of whole exome sequencing (WES) or whole genome sequencing (WGS) to clinical diagnosis, genetic diagnosis of cases with ASDs will further increase the diagnostic yield in the future (15,16).
ASDs are genetically complex disorders. Around 20-30% ASDs patients with a family history of developmental disability or psychiatric problems have multiple genes copy-number variants (CNVs), which can be detected by two different chromosomal microarray (CMA) techniques: array-CGH and single-nucleotide polymorphism arrays (Table 2) (16). Due to the diagnostic yield, CMA is currently used as the first-tier genetic test for individuals with developmental disabilities or congenital anomalies (16).
Other single genetic causes for ASDs include Fragile X mental retardation 1 (FMR1) gene and methyl-CPG-binding protein 2 (MECP2) on the chromosome X and phosphatase and tensin homolog (PTEN) with around 5% detection frequency (Table 2). In those with ASD and head circumferences >2.5 SDs, 5% of them carry PTEN mutations (16).
Fragile X syndrome (FXS) is the most common monogenic cause of developmental cognitive impairment, characterised by intellectual disability and hyperactivity. The average age of FXS diagnosis is 35 to 37 months for boys and 42 months for girls (17). More than one third (37.6%) of families reported that they had more than ten visits before the diagnosis of FXS could be made (17). A trinucleotide CGG repeat in the 5'-untranslated region of the FMR1 gene contains up to 55 repeats of the CGG in the normal population, while in patients with FXS, it can exceed 200 (18,19). Individuals with FXS are at a higher risk of developing seizures (10-40%) and 1-5% patients with autism are due to FMR1 gene abnormality (16).
Rett syndrome is a neurodevelopmental disorder of grey matter of the brain, caused by mutations in the gene of methyl CpG binding protein 2 (MECP2). Most Rett syndrome patients are female raising the possibility that this is a male lethal condition. Clinically, children with Rett syndrome are only noticed after 6-18 months, when they show signs of regression in their speech and motor function capabilities. Growth retardation, cognitive and motor impairment (such as repetitive stereotyped hand movement, dystonia, ataxia, etc.) are often seen. Four precent patients with autism are due to mutations in MECP2 gene (16). Mutations in MECP2 gene could alter the structure or lead to reduced amounts of MeCP2 protein, which binds to methylated DNA and supress gene expression.
Angelman syndrome (AS) & Prader Willi syndrome (PWS): they are the first reported disorder caused by epigenetic imprinting in human. Clinically, AS is characterised by intellectual disability, delayed speech, jerky walking style and happy appearance. PWS is characterised by short stature, skeletal abnormalities, eye problems, intellectual disability and obesity due to excessive eating. The chromosome region responsible for these two disorders are located at chromosome 15q11-13, so called PWS/AS region. For AS, approximately 70% of cases result from de novo deletions of this region on the maternal chromosome, approximately 2% of AS result from paternal uniparental disomy (UPD, meaning both copies of the chromosome inherited from one parent), and 2-3% of AS result from imprinting centre defects. Most of the rest 25% AS are caused by mutations in the gene of ubiquitin-protein ligase E3A (UBE3A). For PWS, approximately 70-80% patients are caused by deletion of this region on the paternal chromosome, the remaining cases result from maternal UPD. DNA-based methylation testing would be a useful diagnostic evaluation for clinically suspect and FISH-negative patients (20).
Adrenoleukodystrophy (ALD, also known as X-linked adrenoleukodystrophy) is caused by mutations in ATP-binding cassette transporter D1 gene (ABCD1), encoding a peroxisomal membrane transporter protein, and its gene mutations result in very-long chain fatty acids accumulation in tissues throughout the body. The most severely affected brain region is white matter containing myelin. Clinical presentation of ALD varies greatly, and the childhood onset type ALD affects only boys. They initially could present with emotional instability, hyperactivity and disruptive behaviour at school. Girls carrying this gene mutation will become symptomatic in adulthood but will be less severe (3).
Genetics of hereditary neuron peripheral disorders
Charcot-Marie-Tooth is also known as hereditary motor and sensory neuropathy (HMSN) and peroneal muscular atrophy (PMA), characterised by patients initial experience of foot drop and gradually muscle tissue wasting of the lower part of the legs (‘inverted bottle’ sign). It is a heterogeneous genetic disorder, caused by mutations in over 50 genes. Most mutations affect the myelin sheath (CMT1, CMT3, CMT4) and some mainly affect the axon (CMT2). CMT1 is the most common genetic cause responsible for 70-80% of all CMT cases due to a duplication of chromosome 17p11.2 involving the PMP22 gene (21) (Table 3).

Full table
Myotonic dystrophy (MD) is an AD disorder characterized by muscle weakness, myotonia, early onset cataracts, and multi-systemic disease. There are two types of MD. An expanded trinucleotide repeats of CTG in the DMPK gene located in chromosome 19 is the cause of MD type I. An expanded CCTG tetranucleotide repeats in intron 1 of zinc finger protein 9 (CNBP) causing reduction of CNBP gene expression and its protein levels is the cause of MD type II. Type II MD is generally milder compared to type I MD. In contrast to Charco-Marie-Tooth, the muscle weakness starts from the hands, neck, face or feet, and slowly progresses to other muscle groups.
Duchenne muscular dystrophy (DMD) & Becker muscular dystrophy (BMS) are X linked disorder, caused by mutations in dystrophin (DMD) gene located in chromosome Xp21.2-p21.3. DMD only affects boys before age 6 with initial symptoms of pelvis associated muscles weakness and progressive proximal muscle dystrophy with the calves initial enlargement (pseudohypertrophy) from ages of 5-15, and great elevation of creatine kinase levels in the blood. As the disease progress, muscle deterioration spreads to other parts of the body, movement lost, and eventually paralyzed. Gower’s sign is a characteristic physical sign of DMD. BMS usually appear in men at about age 8 to 25 with the same symptom development pattern as DMD, but much slower rate of progression. In contrast to DMD, BMD can affect female. The incidence of BMD is about 1 in 17,000 live births, about one fifth of that in DMD (22). About 50% BMD cases have family history and about 38.5% DMD cases are familial cases. Total loss function of dystrophin due to deletions (~60%), duplication (10-15%) and point mutations in the dystrophin leads to DMD and dysfunction or insufficient dystrophin protein due to mutations leads to BMS. Destruction of muscle tissue could be identified by electromyography and positive genetic testing results will ascertain the diagnosis of DMD & BMS.
Spinal muscular atrophies (SMAs) are a group disorder of lower motor neurons degeneration affecting the anterior horn of the spinal cord leading to neuronal death. SMAs are caused by mutations/deletions in multiple genes, such as SMN1, UBA1, ATP7A, IGHMBP2, etc. Clinically, SMAs have various presentations with different disease onset ages and progression (Table 3). The most common gene affected is the deletion of the exons 7 and 8 of the SMN1 gene with a carrier frequency of ~1 in 40 in Caucasians.
Neurofibromatosis (NF) type 1 and tuberous sclerosis complex (TSC) are the two common inherited neurocutaneous disorders in children with different organs initially attacked at different period of age (23,24). Both NF1 and TSC have brain tumour. Gliomas occur frequently in children with NF1 and giant cell astrocytoma is often seen in TSC. Tumours on skin and in other organs are typical feature of TSC. Skin café-au-lait spots could be seen in patients with NF1 or TSC. Deletions and mutations in NF1 gene are the genetic cause of NF1. Deletions and mutations in TSC1 & TSC2 genes, encoding for proteins hamartin and tuberin respectively, are the genetic causes of TSC (Table 3). These genes products have tumor suppress function.
Mitochondrial diseases (MIDs) are a group of systematic disorders. Some MIDs are caused by defects in the mitochondrial genome which is purely inherited from the mother (25). Deficiency in nuclear genes encoding mitochondrial protein is the common cause of MIDs. The characteristic signs of children with MIDs are muscle weakness and loss of muscle coordination. Other clinical features, such as slow growth, learning disabilities, seizures, dementia, and multi-organs dysfunction could also present (Table 3).
Genetics of epilepsy
Inherited epilepsy syndromes can be classified based on their genetic contribution to the aetiology as monogenic epilepsies (caused by defects in genes regulating voltage or ligand-gated ion channels) and complex epilepsies (caused by the interaction of a few or several genetic variants). In addition to the complex genetic contribution, environmental factors also play a role in epilepsy occurrence. Thus, heterogeneous clinical presentations of epilepsy are often seen in clinics.
Genetic generalized epilepsy (GGE) accounts for 30% of all epilepsy (26) and is a genetic complex disorder. Common variants in CHRM3, SCN1A, VRK2, ZEB2 and PNPO have been first identified associating with GGE via genome wide association studies (GWAS) study (27). The identification of genetic risk factors in GGE has been intensely investigated through candidate genes approach, GWAS approach [identifying common variants with minor allele frequency (MAF) >5%] (6,27) or exome sequencing approach (identifying single rare variants of large effect on GGE) (28). Due to its genetic complex disorder by nature, it would not be discussed further in this section.
Familial Mendelian inheritance form of genetic focal epilepsy has been attributed to mutations in the potassium channel genes, such as KCNQ2, KCNQ3 in benign familial neonatal seizures (BFNS), PRRT2 gene (encoding a transmembrane protein and an ion channels regulator) in benign familial infantile seizures (BFIS), and SCN2A gene (encoding sodium channel, α2 subunit) in benign familial neonatal infantile seizures (BFNIS), an intermediate clinical variant of BFIS and BFNIS (Table 1) (26). About 70% BFIS families carry PRRT2 mutations. A total of 80% of patients with Dravet syndrome, a subtype of epileptic encephalopathies, are caused by mutations in SCN1A, representing a relatively genetically homogeneous epilepsy (29). Recently, de novo mutations in genes encoding nicotinergic cholinergic receptors, ion channels and other molecular pathways have been identified causing AD nocturnal frontal lobe epilepsy (ADNFLE), malignant migrating partial seizures in infancy (MMPSI), and epileptic encephalopathies (Table 4) (29-32). Other neurological features could also occur in patients with inherited epilepsy (Table 4).

Full table
Febrile seizures (FS) occur in 2-6% of children between 6 months to 5 years old. Generalised epilepsy with febrile seizers (GEFS) often first occurs in early childhood, and continues after the age of 6 in absence of fever. Seizures stop in mid-adolescence most without neurological developmental abnormalities. Mutations in GABA (A) receptor γ2-subunit gene (GABRG2), and sodium channel genes, encoding the α1, β1 and α2 subunits have been implicated for GEFS (Table 4).
Challenging of genetic testing for paediatric HNDs
One paediatric HND can be caused by mutations in different genes and mutations in the same gene can cause different clinical disorders. For clinicians, a set of clinical phenotypes may suggest a possible diagnosis, and hence a specific gene test will be requested in order to confirm the diagnosis. Not all genes responsible for a certain paediatric HND have been identified. Differentiating true disease-causing mutations from the rare variants is a challenge. In vitro functional testing, animal model or structural analysis would help to establish disease-causing genes. NGS could play a role in pathogenic variant identification and prioritization of candidate variants. Variants which are present in public SNP databases with MAF over 5% shall be excluded, as it is assumed that common variants represent harmless variations. Another way to narrow down the genomic candidate variants is the use of pedigree information. However, a de novo mutation could not be predicted and it is often difficult to show whether a disease follows a recessive or dominant inheritance without family information. The use of different prioritization approaches and the combination of prediction results with phenotypic and pedigree data as well as data from databases might be the best approach to determine the potential cause of the disease under investigation (33).
The same gene mutations causing different clinical phenotypes
Proline-rich transmembrane 2 (PRRT2) causative HNDs: PRRT2 is a small gene of four exons, and it is highly expressed in the basal ganglia and hippocampus. Deletion or mutations in PRRT2 gene causing loss its function leading to paroxysmal kinesigenic dyskinesia was first reported by two independent research groups from China (34,35). Subsequently, mutations in PRRT2 were found in patients with paroxysmal nonkinesigenic dyskinesia (PNKD), paroxysmal exercise-induced dyskinesia (PED) (36), benign infantile convulsions (BFIS), infantile convulsions with choreoathetosis (ICCA), nocturnal convulsions (NC), paroxysmal torticollis and hemiplegic migraine (37), infantile non-convulsive seizures (INCS), febrile convulsions, but not in infantile epileptic encephalopathies (38). Taken together, PRRT2 mutations account for a wide range of different paroxysmal disorders. This provides a challenge on the concept of clinical presentation driven genetic test.
KCNT1 causative HNDs: ADNFLE and malignant migrating partial seizures of infancy (MMPSI) are two completely different epilepsy syndromes, one starts in childhood (ADNFLE) with frontal lobe involvement and the other in infancy (MMPSI) with epileptic encephalopathies. However, mutations in the sodium-gated potassium channel gene (KCNT1) cause both ADNFLE and MMPSI (32,39). This is another example of challenges of current clinical presentation driven genetic test.
Genetic testing strategies and limitations
Genetic testing is a complex area. There is no a single test that can detect all HNDs. The cost of genetic tests is in general high. Therefore accurate clinical diagnosis and careful selection of genetic tests to maximise the detection rate is very important.
Specific gene analysis is to screen mutations in specific genes of interest. For some disorders, the clinical features are well defined and will guide the selection of specific genetic testing, for example, DMD, SMA, HD, etc. It is more economical and effective to test the specific gene as the first line of genetic investigation as the diagnostic yield is expected to be high. Genetic laboratories in different countries offering specific gene testing could be found in the following database (http://www.ncbi.nlm.nih.gov/gtr/; www.genetests.org).
Fragment analysis: tri/tetra nucleotide repeat expansion is responsible for several HNDs, such as fragile X, SCAs, Friedreich’s ataxia, MD, Huntington’s disease, etc. The most common technology used to detect the nucleotide repeats expansion is PCR followed by fragment analysis. The repeat-primed PCR and subsequent fragment separation is capable to detect the presence of the expansion, and also able to distinguish between pure nucleotide repeats and those containing interruptions (40). For large expansion, such as full mutations in fragile X and MD, Southern blotting is often used to estimate the size of nucleotide repeats expansion.
Chromosomal microarray analysis (CMA): although the true mutation spectrum for many HNDs is largely unknown, the use of CMA for the detection of copy number variants (CNVs, single- and multi- exon deletions and duplications) has been successfully applied to children with intellectual disability and/or autism. CNVs account for 7.9% of seizure cases with cognitive impairment or regression (epileptic encephalopathies) (41,42) or 28% cases of movement disorders with neurodevelopmental or behaviour problems (43). Selection of cases with family history and associated comorbid features (such as developmental delay, intellectual disability and behavioural problems) appears to give a significant diagnostic yield and would favour chromosomal microarray as a first-tier investigation for children with intellectual disability or ASDs (44). Multiplex ligand-dependent probe amplification (MLPA) and FISH could be used for further family study of the presence of CNVs (37).
NGS panel sequencing: due to similar clinical presentations can be caused by mutations in different genes, majority of HNDs could not be clearly diagnosed in clinical practice. Panels of multiple genes responsible for a certain disorder spectrum can be designed and developed to target a spectrum of HNDs, such as movement disorders, epilepsy, developmental and neuron peripheral disorders, as listed above. The NGS technology provides a powerful platform to sequence multiple genes simultaneously.
WES or WGS: genome wide tests have significantly advanced the discovery of novel causative genes for HND (28,34,35). The clinical diagnostic application of NGS technologies is promising but challenging (34). Distinguishing the causative from non-causative variations is difficult with the increasing amount of data generated through massive parallel sequencing studies (29). Rare genetic variants, which are expected to contribute significantly to HNDs, are still difficult to identify. As exome sequencing enters the mainstream in clinical practice, sharing of research data, central repositories for clinical data and genetic data will become essential resources for interpreting NGS data to identify the disease causative mutations (29).
Others: karyotype and FISH analysis have been used as confirmatory tests after the screening tests. Due to their limitations in detecting point mutations and small genetic changes, they are not considered as the first line investigation for patients with neurological disorders. Although AS and PWS are imprinting disorders, DNA methylation analysis is not considered as the first line investigation, as majority of AS and PWS are caused by deletions.
Ethics of genetic testing
In paediatric HNDs practice, the most common scenario is diagnostic and pre-symptomatic genetic testing, although genetic screening, carrier testing, etc., may also be conducted. Genetic information is complex and needs to be presented by a genetic counsellor in a meaningful and non-judgmental way that will benefit the patients and their families in terms of decision-making, personal control and minimizing psychological distress. Knowing the genetic cause may help to understand the prognosis of the disorder as well as assessing the recurrence risk within a family. Support services and therapeutic reagents may also be more easily obtained with a specific genetic diagnosis (45).
Diagnostic genetic testing: a written consent from a responsible parent shall be obtained before the genetic test. Sometimes, if older children show adequate competence in the decision-making, both the parents and child might choose to sign the consent form (46,47). In some situations, pre-test counselling could be replaced by pre-test information about the reasons to undertake the test and the implications involved (48). The issues of how genetic counselling is affected by the appropriateness, choice of protocols, interpretation of results, and ethical and social concerns of the genetic tests for HNDs in children has been addressed well in a review by Valente et al. (48). If it is a positive finding, genetic anticipation, incomplete penetrance and sexual phenotype differences shall be discussed, particular in those disorders caused by genes CNVs or tri/tetra nucleotide repeat expansion or X-linked (48). If it is a negative result, the putative diagnosis, screening techniques and sensitivity factors shall be excluded and defined as true negative (48). In term of the diagnostic application of whole genome and/or WES, the incidental or unrelated findings from the initial genetic testing purpose, such as undisclosed relationships and adult onset cancer susceptibility, etc., should not be disclosed without consent (45).
Pre-symptomatic genetic testing: it refers to children and young people who have a family history of a heritable condition but do not have symptoms or signs of the condition and the children are almost certain to develop the condition during their life time, such as HD, if carrying the mutated gene. There are several guidelines regarding whether and when to do pre-symptomatic genetic testing in children and young adults (49): (I) pre-symptomatic genetic testing should be offered, if there is potential medical benefit in the immediate future, such as Wilson disease; (II) pre-symptomatic genetic testing should not be undertaken if there is no medical benefit in the immediate future, such as HD; (III) if a condition that always has its onset before adolescence, such as DMD, and the child is too young to make the decision, parents may have the right to request a genetic test for an asymptomatic at-risk sibling; (IV) if a condition can have its onset at an age in adolescence or adulthood, it is recommended that the genetic test should not be done until the children are matured enough to be able to involve in the decision making for genetic testing.
Future directions of the field
Genetic testing on paediatric neurological disorders is a translational discipline from research based technological progresses to the clinics. It is a multifaceted process as discussed in this review. Genetic counselling, the appropriateness and quality of the test, the credibility of the provider, and the most important information of the context of the patient’s symptoms and family history are essential for the interpretation of genetic testing results and for the best management of patients with neuro paediatric disorders and their families (48). Clinical physical examination, neuroimaging and biochemistry tests would guide one’s selection of genetic testing to increase the positive yield (45).
As new genomic technologies offer insight into previously undiagnosed neurologic disorders, we will discover new syndromes and expand the disease spectrum of different disorders caused by a single gene. Identifying HNDs causative genes is critical for the development of targeted treatments and prevention approach. This review equips paediatricians to better handle the challenges during the upcoming genomic era.
Acknowledgements
Funding: This work is supported by Australian national health and medical research (NHMRC) research program (1037746), and the State Key Program of the National Natural Science Foundation of China (81130021).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Williams K, Thomson D, Seto I, et al. Standard 6: age groups for pediatric trials. Pediatrics 2012;129:S153-60. [PubMed]
- Contopoulos-Ioannidis DG, Seto I, Hamm MP, et al. Empirical evaluation of age groups and age-subgroup analyses in pediatric randomized trials and pediatric meta-analyses. Pediatrics 2012;129:S161-84. [PubMed]
- Piña-Garza JE. eds. Fenichel’s Clinical Pediatric Neurology: A Signs and Symptoms Approach (Expert Consult - Online and Print), 7e. Elsevier, 2013.
- Chen G, Shi T. Next-generation sequencing technologies for personalized medicine: promising but challenging. Sci China Life Sci 2013;56:101-3. [PubMed]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med 2010;61:437-55. [PubMed]
- Petrovski S, Kwan P. Unraveling the genetics of common epilepsies: approaches, platforms, and caveats. Epilepsy Behav 2013;26:229-33. [PubMed]
- Charlesworth G, Bhatia KP, Wood NW. The genetics of dystonia: new twists in an old tale. Brain 2013;136:2017-37. [PubMed]
- Fujioka S, Sundal C, Wszolek ZK. Autosomal dominant cerebellar ataxia type III: a review of the phenotypic and genotypic characteristics. Orphanet J Rare Dis 2013;8:14. [PubMed]
- Gasser T, Finsterer J, Baets J, et al. EFNS guidelines on the molecular diagnosis of ataxias and spastic paraplegias. Eur J Neurol 2010;17:179-88. [PubMed]
- Bhidayasiri R, Pulst SM. Juvenile parkinsonism. Eur J Paediatr Neurol 2009;13:290-2. [PubMed]
- Albanese A, Bhatia K, Bressman SB, et al. Phenomenology and classification of dystonia: a consensus update. Mov Disord 2013;28:863-73. [PubMed]
- Quarrell O, O’Donovan KL, Bandmann O, et al. The prevalence of juvenile Huntington’s disease: a review of the literature and meta-analysis. PLoS Curr 2012;4:e4f8606b742ef3.
- Scuffham TM, Macmillan JC. Huntington disease: who seeks presymptomatic genetic testing, why and what are the outcomes? J Genet Couns 2014. [Epub ahead of print]. [PubMed]
- Filocamo M, Morrone A. Lysosomal storage disorders: molecular basis and laboratory testing. Hum Genomics 2011;5:156-69. [PubMed]
- Hindley D, Medakkar S. Diagnosis of Down’s syndrome in neonates. Arch Dis Child Fetal Neonatal Ed 2002;87:F220-1. [PubMed]
- Schaefer GB, Mendelsohn NJ; Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med 2013;15:399-407. [PubMed]
- Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012;485:237-41. [PubMed]
- Yu S, Pritchard M, Kremer E, et al. Fragile X genotype characterized by an unstable region of DNA. Science 1991;252:1179-81. [PubMed]
- Yu S, Mangelsdorf M, Hewett D, et al. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 1997;88:367-74. [PubMed]
- Velinov M, Gu H, Shah K, et al. PCR-based methylation testing for Prader-Willi or Angelman syndromes using archived fixed-cell suspensions. Genet Test 2001;5:153-5. [PubMed]
- Raeymaekers P, Timmerman V, Nelis E, et al. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord 1991;1:93-7. [PubMed]
- Manzur AY, Muntoni F. Diagnosis and new treatments in muscular dystrophies. J Neurol Neurosurg Psychiatry 2009;80:706-14. [PubMed]
- Roach ES, Sparagana SP. Diagnosis of tuberous sclerosis complex. J Child Neurol 2004;19:643-9. [PubMed]
- DeBella K, Szudek J, Friedman JM. Use of the national institutes of health criteria for diagnosis of neurofibromatosis 1 in children. Pediatrics 2000;105:608-14. [PubMed]
- Falk MJ, Sondheimer N. Mitochondrial genetic diseases. Curr Opin Pediatr 2010;22:711-6. [PubMed]
- Perkin GD, Johnson MR. Epilepsy in later childhood and adulthood. In: Warrell DA, Cox TM, Firth JD. eds. Oxford Textbook of Medicine (5 ed.) Oxford: University Press, 2013.
- EPICURE Consortium; EMINet Consortium, Steffens M, et al. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Hum Mol Genet 2012;21:5359-72. [PubMed]
- Heinzen EL, Depondt C, Cavalleri GL, et al. Exome sequencing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet 2012;91:293-302. [PubMed]
- Helbig I, Lowenstein DH. Genetics of the epilepsies: where are we and where are we going? Curr Opin Neurol 2013;26:179-85. [PubMed]
- Schmidt A, Kumar KR, Redyk K, et al. Two faces of the same coin: benign familial infantile seizures and paroxysmal kinesigenic dyskinesia caused by PRRT2 mutations. Arch Neurol 2012;69:668-70. [PubMed]
- Depienne C, Gourfinkel-An I, Baulac S, et al. Genes in infantile epileptic encephalopathies. In: Noebels JL, Avoli M, Rogawski MA, et al. eds. Jasper’s Basic Mechanisms of the Epilepsies [Internet]. 4th edition. Bethesda (MD): National Center for Biotechnology Information (US), 2012.
- Barcia G, Fleming MR, Deligniere A, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet 2012;44:1255-9. [PubMed]
- Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 2014;15:256-78. [PubMed]
- Wang JL, Cao L, Li XH, et al. Identification of PRRT2 as the causative gene of paroxysmal kinesigenic dyskinesias. Brain 2011;134:3493-501. [PubMed]
- Chen WJ, Lin Y, Xiong ZQ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet 2011;43:1252-5. [PubMed]
- Wang JL, Mao X, Hu ZM, et al. Mutation analysis of PRRT2 in two Chinese BFIS families and nomenclature of PRRT2 related paroxysmal diseases. Neurosci Lett 2013;552:40-5. [PubMed]
- Dale RC, Gardiner A, Antony J, et al. Familial PRRT2 mutation with heterogeneous paroxysmal disorders including paroxysmal torticollis and hemiplegic migraine. Dev Med Child Neurol 2012;54:958-60. [PubMed]
- Liu XR, Wu M, He N, et al. Novel PRRT2 mutations in paroxysmal dyskinesia patients with variant inheritance and phenotypes. Genes Brain Behav 2013;12:234-40. [PubMed]
- Heron SE, Smith KR, Bahlo M, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet 2012;44:1188-90. [PubMed]
- Holloway TP, Rowley SM, Delatycki MB, et al. Detection of interruptions in the GAA trinucleotide repeat expansion in the FXN gene of Friedreich ataxia. Biotechniques 2011;50:182-6. [PubMed]
- Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol 2011;70:974-85. [PubMed]
- Nicholl J, Waters W, Suwalski S, et al. Epilepsy with cognitive deficit and autism spectrum disorders: prospective diagnosis by array CGH. Am J Med Genet B Neuropsychiatr Genet 2013;162B:24-35. [PubMed]
- Dale RC, Grattan-Smith P, Nicholson M, et al. Microdeletions detected using chromosome microarray in children with suspected genetic movement disorders: a single-centre study. Dev Med Child Neurol 2012;54:618-23. [PubMed]
- Kurian MA. The clinical utility of chromosomal microarray in childhood neurological disorders. Dev Med Child Neurol 2012;54:582-3. [PubMed]
- Vento JM, Schmidt JL. Genetic testing in child neurology. Semin Pediatr Neurol 2012;19:167-72. [PubMed]
- Davis DS. Genetic dilemmas and the child’s right to an open future. Hastings Cent Rep 1997;27:7-15. [PubMed]
- Darby RJ. The child’s right to an open future: is the principle applicable to non-therapeutic circumcision? J Med Ethics 2013;39:463-8. [PubMed]
- Valente EM, Ferraris A, Dallapiccola B. Genetic testing for paediatric neurological disorders. Lancet Neurol 2008;7:1113-26. [PubMed]
- Human Genetics Society of Australia. Pre-symptomatic testing in children and young adults. 2008;2008PS02.


