Fanconi anemia pathway defects in inherited and sporadic cancers
Introduction
Fanconi anemia (FA) was first described and named in 1927 by the Swiss paediatrician Guido Fanconi who treated three brothers suffering from aplastic anemia and congenital malformations (1). So far, 16 different FA causing genes have been identified and such advancing studies on the FA pathways increased the understanding of both the phenotype and genotype of the disease condition.
Studies on the mechanism of FA demonstrated that the mutations in cancer genes are involved in repairing DNA, regulating replication-dependent removal of interstrand DNA crosslinks (ICLs) and maintaining of genomic stability affecting many cellular processes such as repairing DNA, regulating replication-dependent removal of ICLs. Therefore, it is not surprising that FA is associated with increased risk in developing multiple types of cancer (2).
The cause of cancers is diverse and complex. They are associated with both genetic (familial inherited) and environmental (acquired) risk factors. It is becoming evident that there is strong link between FA and cancers from recent studies (3). Studies on the link between cancer from bi-allelic and mono-allelic FA genes mutated provide a very promising direction in cancer research. Substantial results obtained from FA studies indicate that FA is the suitable model in cancer studies because cancer from FA gene mutations is associated with both genetic and environmental factors. Understanding the connection between FA genes and cancer genes potentially holds the key for cancer research into the mechanisms of susceptibility and suppression, as well as clinical therapies.
Identification of FA genes
FA is classified into 16 different complementation groups. The 16 different corresponding genes have been identified and named so far and each subtype of these 16 genes is linked to a different disease gene (Table 1). It is still possible to find more FA genes mutated related to the FA phenotype. Fifteen of 16 FA genes are inherited in an autosomal recessive manner but only the subtype, FANCB is in the sex chromosome and inherited in an X-linked manner (2-4).
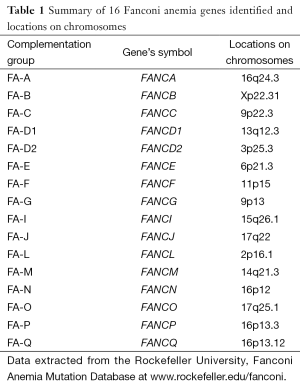
Full table
In the general population, the complementation-A occurs in about 60-70%, while complementation-C occurs in about 15% and the complementation-G occurs in about 10% of the total 16 FA responsible genes mutated with a vary of their subgroups in some geographical regions such as in Mediterranean countries (5). About 80% FA patients belong to this complementation group A in Spain (6).
Molecular studies of FA pathway defects in cancers
The FA pathways are defined from the encoded proteins work in concert in a distinct genomic maintenance. The FA pathways in normal cells are not constitutionally active but they are turned on during the S phase of the cell cycle in the presence of DNA damage together to coordinate distinct repair functions in nucleotide excision, translesion synthesis and homologous recombination to remove the cross links (7).
The 16 FA gene products make up the FA pathways in the maintenance of genomic stability and the FA pathways can be activated by DNA damage and replication. The number of FA proteins reflects the complicated nature of the FA pathways. Mutation in any of the 16 FA genes cause defects in the response to DNA damage and repair and result in disease of FA by the loss of DNA interstrand crosslinks repair (8). The FA pathways are the key event with the complex pathological mechanisms in DNA repair and cancer suppression in both inherited and sporadic cancers.
The proteins involved in FA consist of several classes of enzymes and structural proteins including ubiquitin ligase, monoubiquininated proteins and helicase (9). FA nuclear core protein complex consists of eight proteins, encoded by FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM, with ubiquitin ligase activity. This protein complex is required for the critical monoubiquitination of FANCD2 and FANCI in response to DNA damage during replication. The biological functions of this protein complex are to maintain DNA stability and repair DNA damage Protein-protein interactions are required for core complex protein stability form the stable core protein complex in function that is required for the modification of FANCD2 and FANCI by monoubiquitination.
E3 ubiquitin-protein ligase FANCL belongs to the multisubunit FA complex and it is a ligase protein that mediates monoubiquitination of FANCD2, a key step in the repair of ICLs in the FA pathways (10). FANCL is associated with hypersensitivity to DNA-damaging agents, chromosomal instability (increased chromosome breakage) and defective DNA repair (11).
The monoubiquitination process of FANCD2 and FANCI recruits DNA repair machinery in order to maintain genomic integrity during cellular proliferation within certain tissues. Mutation in any member of a core protein complex result in the loss of the monoubiquitination of the FANCI/FANCD2 complex step (12). FANCD2 and FANCI proteins are substrates for ubiquitination with the two being similar in size and domain structure. This monoubiquitination is the crucial event of the FA pathways, and the monoubiquitinationed isoform of FANCD2 associates with the repair protein BRCA1 in DNA damage induced nuclear foci. This foci formation is induced by both crosslinking agents and DNA damaging agents (13). This process is regulated by the nuclear core protein complex.
Patients with FA from homozygous mutation increase cancer susceptibility
Inherited homozygous (bi-allelic) mutations from germ line can cause FA and increase susceptibility to both hematologic and non-hematologic malignancies (14). The first case of FA was recognised as a cause of Juvenile leukemia in 1967 (15). A close connection between FA and tumorigenesis has been observed from both clinical and cellular phenotypes. Cancers with FA gene mutations are difficult to be treated (except surgically) because cells from patients with FA are more sensitive to chemotherapy and radiation comparing with non-FA cancers (16). The risk of developing to acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) has been reported to increase by 785 fold (17,18).
The relative risk of non-hematologic malignancies in patients with FA is increased commonly for squamous cell carcinomas (700 times greater than in normal population) including the head and neck, vulva, esophagus, gastric osteogenic sarcoma, cervix and skin. Many other types of cancers including breast cancer, lung cancer, colon cancer, brain tumour were found from patients with FA as well, with the median onset age of cancers being 16 years old in FA patients compared with 68 years in the non-FA population (19,20). In addition, FA patients may have a combination of different types of cancers.
Heterozygous FA gene mutation increase sporadic cancers in non-FA patients
The genetic association between cancers with FA and without FA mutation clinically has been intensively investigated. Heterozygous FA gene (mono-allelic) mutations somatically do not cause FA phenotype but significantly increase cancer susceptibility in a sporadic manner. Sporadic cancer is the term commonly used to describe malignancies occurring in individuals at relatively old age and without an observable family history of cancer. In other words, sporadic cancer is caused mainly by environmental factors. Studies on four FA genes (FANCD1, FANCJ, FANCN and FANCO) of the 16 identified from patients with FA strengthened the association between FA and breast cancer and FANCD1 was found to be identical to BRCA2 and the active isoform of FANCD2 interacted with BRCA1 (20-26).
Mutations in FANCJ and FANCN have also been found to cause both FA and cancer (when bi-allelic) but such mutations increased breast cancer risk but not FA (when mono-allelic) (27). Similarly, homozygous missense mutation of FANCO (RAD51C),a newly identified cause of FA found from a consanguineous family with three affected children showed congenital anomalies characteristic of FA and also monoallelic mutations associated with risk of breast and ovarian cancer was reported (28) and a separate study reported that sporadic pancreatic cancer from the FANCC gene mutation and the FANCG gene mutation both with the loss of heterozygosite (29). Studies also showed that mono-allelic FA gene mutation can contribute to the pathogenesis of several types of cancers from the consequence of haplo-insufficiency as the carcinogenic mechanism in the different penetration degree with the addition of second hit typically seen in colorectal cancer from MLH gene mutation and in breast cancer from BLM gene mutation (30).
The accumulated evidence suggests that epigenetic changes in FA genes also play a role in sporadic cancer in the general population. The FA proteins inhibit cancer cells by promoter hypermethylation. There is a strong link between FA and cancer pathogenesis by affecting DNA repair, regulation pathway and epigenetic mechanism (31). Silencing of some FA genes were observed such as FANCF, FANCL, FANCC, FANCB, BRCA2/FANCD1 and FANCN/PALB2 inhibition in cancer cells by a promoter hypermethylation process in several types of cancers without FA phenotype. Studies showed that pathogenic mechanisms involved in disruption of FA pathways in sporadic cancer were in epigenetic silencing of FA genes, somatic mutations leading to loss of function of FA genes and elevated expression of silencing variants in the aspects of down-regulation and competition with functional FA proteins. A study also showed that hypermethylation of FANCF is the most frequent epigenetic change leading to FA pathway disruption in different cancer types including AML, ovarian cancer and cervical cancer, head and neck squamous cell cancer, non-small cell lung cancer, granulose cell tumours, breast and bladder cancer (32,33).
The important difference between familial/inherited and sporadic cancer is due to the form of germ line mutation in a DNA caretaker gene facilitating the accumulation of oncogenic DNA changes which it can result in a high susceptibility of cancer. It is clear that sporadic mono-allelic FA gene mutation leads to increased cancer risk but study also suggests that the familial mono-allelic FA gene mutated (familial carrier) does not show a high cancer risk than normal population or due to the size of cases numbers studied.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Fanconi G. Familiäre infantile perniziosaartige Anämie (perniziöses Blutbild und Konstitution). Jahb Kinderheilkd 1927;117:257-80.
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A 2009;106:7155-60. [PubMed]
- Osorio A, Bogliolo M, Fernández V, et al. Evaluation of rare variants in the new fanconi anemia gene ERCC4 (FANCQ) as familial breast/ovarian cancer susceptibility alleles. Hum Mutat 2013;34:1615-8. [PubMed]
- Strathdee CA, Duncan AM, Buchwald M. Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet 1992;1:196-8. [PubMed]
- Tipping AJ, Pearson T, Morgan NV, et al. Molecular and genealogical evidence for a founder effect in Fanconi anemia families of the Afrikaner population of South Africa. Proc Natl Acad Sci U S A 2001;98:5734-9. [PubMed]
- Castella M, Pujol R, Callén E, et al. Chromosome fragility in patients with Fanconi anaemia: diagnostic implications and clinical impact. J Med Genet 2011;48:242-50. [PubMed]
- Alpi A, Langevin F, Mosedale G, et al. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol 2007;27:8421-30. [PubMed]
- Akkari YM, Bateman RL, Reifsteck CA, et al. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol 2000;20:8283-9. [PubMed]
- Wang W. A major switch for the Fanconi anemia DNA damage-response pathway. Nat Struct Mol Biol 2008;15:1128-30. [PubMed]
- Hodson C, Walden H. Towards a molecular understanding of the fanconi anemia core complex. Anemia 2012;2012:926787.
- Hodson C, Cole AR, Lewis LP, et al. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem 2011;286:32628-37. [PubMed]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet 2009;43:223-49. [PubMed]
- Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002;297:606-9. [PubMed]
- Stecklein SR, Jensen RA. Identifying and exploiting defects in the Fanconi anemia/BRCA pathway in oncology. Transl Res 2012;160:178-97. [PubMed]
- Fanconi G. Familial constitutional panmyelocytopathy, Fanconi’s anemia (F.A.). I. Clinical aspects. Semin Hematol 1967;4:233-40. [PubMed]
- Eiler ME, Frohnmayer D, Larsen K, et al. eds. Fanconi Anemia, Guidelines for Diagnosis and Management. 3rd ed. Eugene: Fanconi Anemia Research Fund, Inc., 2008:18.
- Alter BP. Inherited bone marrow failure syndromes. In: Nathan DG, Orkin SH, Look AT, et al. eds. Nathan and Oski’s Hematology of Infancy and Childhood. 6th ed. Philadelphia: WB Saunders, 2003:280-365.
- Young NS, Alter BP. eds. Aplastic Anemia: Acquired and Inherited. Philadelphia: WB Saunders, 1994.
- Alter BP. Cancer in Fanconi anemia, 1927-2001. Cancer 2003;97:425-40. [PubMed]
- Kutler DI, Singh B, Satagopan J, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 2003;101:1249-56. [PubMed]
- Rosenberg PS, Alter BP, Ebell W. Cancer risks in Fanconi anemia: findings from the German Fanconi Anemia Registry. Haematologica 2008;93:511-7. [PubMed]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 2002;100:2414-20. [PubMed]
- Zhang F, Ma J, Wu J, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 2009;19:524-9. [PubMed]
- Levy-Lahad E. Fanconi anemia and breast cancer susceptibility meet again. Nat Genet 2010;42:368-9. [PubMed]
- Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410-4. [PubMed]
- Shimamura A, Montes de Oca R, Svenson JL, et al. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood 2002;100:4649-54. [PubMed]
- Goldberg JI, Borgen PI. Breast cancer susceptibility testing: past, present and future. Expert Rev Anticancer Ther 2006;6:1205-14. [PubMed]
- Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 2010;42:406-9. [PubMed]
- van der Heijden MS, Yeo CJ, Hruban RH, et al. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res 2003;63:2585-8. [PubMed]
- Bogdanova N, Helbig S, Dörk T. Hereditary breast cancer: ever more pieces to the polygenic puzzle. Hered Cancer Clin Pract 2013;11:12. [PubMed]
- Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 2006;107:4223-33. [PubMed]
- Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 2001;7:249-62. [PubMed]
- Tischkowitz M, Ameziane N, Waisfisz Q, et al. Bi-allelic silencing of the Fanconi anaemia gene FANCF in acute myeloid leukaemia. Br J Haematol 2003;123:469-71. [PubMed]


