
Predicting the postoperative blood coagulation state of children with congenital heart disease by machine learning based on real-world data
Introduction
In recent years, the ranges of disease entities, biomarkers, diagnostic testing, and treatment modalities all have become increasingly complex and increased exponentially. Subsequently, clinical decision-making has also become more complex and demands the synthesis of decisions from assessment in the current digital age, while the electronic health record represents a massive repository of electronic data points representing a diverse array of medical information (1-3). Now, artificial intelligence (AI) methods have emerged as powerful tools to transform medical care (4) and mine medical data to aid in disease diagnosis and management and perhaps even augmenting, the clinical decision-making of doctors.
AI involves the development of algorithms to perform tasks typically associated with human intelligence (5). Machine learning (ML) is a subfield of AI, has rapidly developed and likely brought changes to current clinical medicine practice (6,7). For cardiology (8) and prostate cancer (9,10), most ML algorithms are viewed as mathematical models that map a set of observed variables (i.e., features, indicators or predictors) into a set of outcome variables (i.e., targets). The outcomes inferred from the expansion of the original scope of clinical data for study, therefore do not necessarily reveal the optimum pathways in terms of efficacy and effectiveness for real-world populations (11). The availability of big data of biomedicine (12,13) together with improvements in data mining, analytics, and ML modeling have sparked an increasing number of real-world evidence studies (14) and have facilitated personalized and outcome-based health care (15). Razavian et al. and colleagues have shown the benefit of using clinical data to develop predictive models of diabetes (16), which demonstrated a highly prevalent disorder of glucose metabolism.
According to Williams et al.’s study, the congenital heart disease (CHD) is one of the most common birth defects (17) that affects approximately 1% of infants born each year (18). In the last few years, the fields of pediatric CHD have experienced considerable progress with advances in new therapeutic and diagnostic techniques that can be applied at all stages of life, which from the fetus to the adult. Simultaneously, surgery is also advancing. Abnormal coagulation may occur in children with CHD after surgery, which often has a serious adverse impact on the prognosis and survival rate of children with CHD. In the case of limited resources, a fundamental problem we must face is how to predict postoperative coagulation abnormalities in CHD children more easily and quickly before surgery to implement early clinical intervention and greatly reduce the risk of postoperative coagulation abnormalities in children with CHD.
Screening, as the primary means of preventing secondary coagulopathy, is a regular examination involving specific detection methods for children with CHD before an operation, to alleviate the disability caused by blood coagulation diseases. Our study attempts to use AI (ML) to explore and find more sensitive indicators and a combination of indicators to distinguish an abnormal blood coagulation (configuration) state with CHD children from widely used medical laboratory indicators derived from examination data, such as conventional coagulation tests (CCTs) and complete blood count (CBC) combined with demographic characteristics, which provides better support for guiding blood transfusion or preventing abnormal blood coagulation (configuration) in CHD children. In summary, our study provides proof of concept for implementing an ML-based system as a means to help doctors in tackling large quantities of data, augment diagnostic evaluations, and provide clinical decision support.
We present the following article in accordance with the TRIPOD Checklist: Prediction Model Development (available at http://dx.doi.org/10.21037/tp-20-238).
Methods
Patient selection and characteristics
In our study, the 91,044 pediatric patients in Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health, China, from 1/1/2013 to 9/30/2018 were collected, including multidimensional clinical information such as demographic characteristics of patients, admission information, CCTs and CBC information, and a total of 1,690 patients fulfilled the inclusion criteria, having the same normal blood coagulation function as CHD children before cardiac surgery. Cohort characteristics of children with CHD are presented in Table S1. Clinical data were recorded on pre-specified forms for all patients. Case report form definitions and electronic health records were centrally predetermined. Admission and discharge diagnoses were determined by the attending doctors based on clinical, electrocardiographic, hematologic, and biochemical criteria. Patient management was at the discretion of the attending doctors.
The mean age of the selected children with CHD was 1.751±2.731 years, and 55.680% (941/1,690) were males. The majority had congenital pulmonary stenosis, congenital transposition of great arteries, ventricular septal defect, congenital pulmonary stenosis, congenital atrial septal defect, tetralogy of Fallot, congenital anomalous origin of pulmonary artery, congenital pulmonary atresia, pulmonary artery occlusion and residual shunt after repair of atrial septal defect. A total of 703 cases showed abnormal blood coagulation, and 987 cases were normal after cardiac surgery. These data can be used to build the prediction models of postoperative abnormal or normal blood coagulation (observation or control group) in children with CHD.
The CCTs and CBC data were collected during the preoperative 48 hours to postoperative 24 hours, and the indicators without valid data were removed; the list is shown as follows. CCTs include prothrombin time (PT), activated partial thromboplastin time (APTT), international standardized ratio (INR), fibrinogen (FBG), D-dimer, anticoagulant enzyme III (ATIII), INR derived from PT and the International Sensitivity Index (ISI) (19) of the assay reagent. Parameters of CCTs and their reference ranges are in Table S2. In the study, if there was one abnormality indicator in the CCTs, the blood coagulation of the patient was defined as abnormal coagulation.
Fasted blood was sampled, and CBC, including red blood cell (RBC) count, hemoglobin (HB), hematocrit (HCT), platelet count (PLT) and white blood cell count (WBC), mean corpuscular volume (MCV), mean corpuscular hemoglobin concentration (MCHC), and mean corpuscular hemoglobin (MCH), etc. was measured by an automated hematological analyzer Sysmex Xs-800i (Sysmex corporation, Kobe, Japan).
ML algorithms
In the study, five representative supervised classification ML algorithms were selected. We used three prediction models, Decision tree, Naive Bayes, and Support Vector Machine (SVM), to build a prediction model based on the different combinations of variables described above. We also chose a tree-based ensemble classification algorithm (Random Forest) and Adaptive Boost (AdaBoost) to build models based on the combination of the aforementioned variables. Decision tree and Naive Bayes produce models with interpretable structures, whereas Random Forest, AdaBoost and SVM are “black box” models, where the function connecting the predictor variables with response is opaque to the user (20-26). Predictive performance as previously described (27), was assessed by the area under the receiver operating characteristic (ROC) curve [AUC (28)], calibration curve and the Lift curve (30% of the original cohort, randomly selected samples). The performance of ensemble models was compared.
Feature selection and model construction
Features were selected by applying Minimum Exponential Description Length (MEDL) algorithm, which was derived from the MEDL principle. Features were also selected by recursive feature elimination (RFE) as previously described (29). In brief, recursively considering increasingly smaller feature sets. In the study, we chose the RFE cross-validation algorithm (30), which performed the RFE algorithm in the cross-validation cycle, to identify the optimal number of important indicators. Then, ML models (SVM, random forest classifier, etc.) were trained using 70% training set and 30% test set (31). The detailed process of features and model selection is shown in Figure 1.
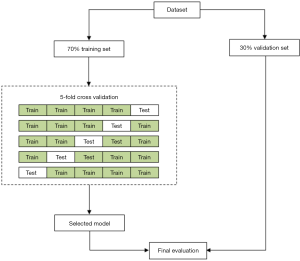
After preoperative CCTs and CBC test data, demographic characteristics data were selected to build the Decision tree classifier model, SVM classifier model, Naive Bayes model, AdaBoost model or Random Forest classifier. The ML algorithms were implemented using Python 3.7.4 (https://www.python.org) with scikit-learn (https://scikit-learn.org/stable/).
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Beijing Children’s Hospital, Capital Medical University gave expedited approval to review and use the medical data, such as electronic health records (ID: 2018-126). The pediatric patients or their guardians were not required to provide written informed consent for our study because of the retrospective nature of the study. The authors have no other ethical conflicts to disclose.
Statistical analysis
The results were expressed as the mean ± standard deviation (SD) for parametric variables and as frequencies/percentages for nonparametric and categorical variables. Differences between groups were analyzed using the Mann-Whitney U test. All analyses were performed using Stata/IC version 16.0 (College Station, TX, Stata Corp. LLC.) or R version 3.6.1 (R Foundation for Statistical Computing). Statistical significance was considered when P values were <0.05.
Results
Primary objective prediction
Important feature for model building
Based on the analysis, we concluded that the verification results of the data mining algorithms were basically consistent with the statistical description results. The observation group and the control group had certain similarities in the distribution. Age and sex play key roles in the construction of ML classification models, even after the completion of a more accurate data selection process for age and sex. Therefore, age and sex would be included in subsequent ML classification models as predictors. In addition, some important indicators were selected, such as MCV, MCH, and MCHC. In summary, five features were selected as important predictors on the basis of data-driven and medical selection, as shown in Table 1.
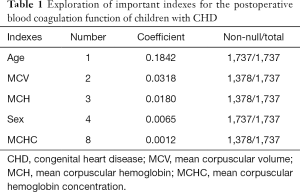
Full table
Classification modeling evaluation
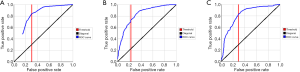
It can be seen from the statistical data that the AUC values obtained by the prediction model are relatively high, indicating that the selected indicators are more effective. The AUCs of the three models were 0.81, 0.82, and 0.82, respectively, which were very close and showed impressive performance. Among them, the true positive rates (TPRs) of both the Decision tree and the SVM prediction models reached more than 80%. For model selection, the AUC was a general metric of performance. However, AUC was not the unique indicator. Their overall accuracies, which were 75.95%, 75.63%, and 75.79%, also indicated that the result was highly effective. Among them, the Decision Tree model was selected to verify the model reliability because it had the best TPR. Further improvements of the classification model need to be achieved for a better prediction result. The specific results are shown in Table 2 and Figure 2.

Full table
Improved modeling for further prediction
Exploration of important features
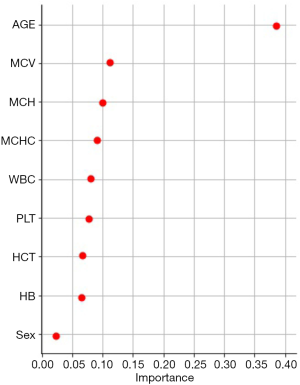
The Decision tree-based algorithm was used to explore the important features related to postoperative blood coagulation of children with CHD. The variable exploration results were as follows (Figure 3). A total of 7 important features were selected as important predictors for modeling on the basis of data-driven and medical selection, the same as primary objective prediction, as shown in Table 3. The establishment of prediction models for predictors can be roughly divided into two aspects: patient attribute characteristics: age, sex; patient-specific test features: MCV, MCH, MCHC, WBC, and PLT.
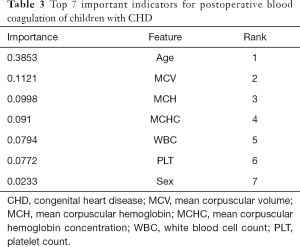
Full table
ML models building
Based on the data processing in the above section, different types of supervised learning algorithms were used to explore the prediction model. The seven important features were included in the learning first. The target variable “postoperative abnormal blood coagulation” was the outcome indicator. Therefore, ML was a prediction model based on multidimensional features, which has better clinical application values. Finally, the three best prediction models were selected, and the ROC curve results are shown in Figure 4. The AUCs of the AdaBoost model, Random Forest model and SVM model are 0.8405, 0.8406 and 0.8387, respectively.

Evaluation of ML models
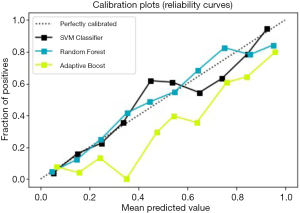
For the three prediction models above, the method based on the calibration curve was used for further evaluation. The calibration curve is a scatter plot of the actual incidence and the predicted incidence. The results are shown in Figure 5. The SVM model is black, the Random Forest model is blue, and the AdaBoost model is yellow; the SVM model and Random Forest model fit better than the others.
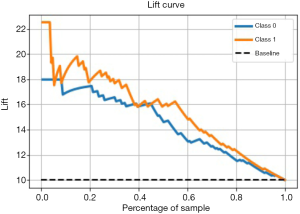
Performance evaluation of excellent models with Lift
The Lift curve is one of the most commonly used methods for ML classification. Lift reflects how many times the accuracy of prediction improves compared to random selection or inference (naive prediction) without using a prediction model. Lift reveals the effect of the prediction model. Unlike the ROC curve, the Lift curve is convex toward the [0, 1] point, and we want to obtain the largest Lift [>1], that is to say, the right half of this curve should be as steep as possible. To obtain a more reliable evaluation of the performance of the prediction model, this study will comprehensively apply ROC curves and Lift curves to verify the performance of the model based on different algorithms. The larger the Lift value of the prediction model is for the SVM model, the better the model effect. As shown in Figure 6, for children with CHD, the SVM model predicted postoperative abnormal blood coagulation, Lift =2.2, which is 2.2 times more accurate than simple prediction, and postoperative normal blood coagulation, Lift =1.8, which is 1.8 times more accurate than simple prediction. The Lift curve basically shows a downward trend, also suggesting that the SVM model has good prediction performance.
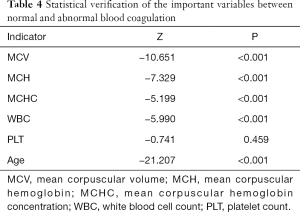
Statistical evaluations with test differences
Statistical tests were performed to distinguish the differences between the observation group (abnormal blood coagulation) and the control group (normal blood coagulation) on variables to verify the conclusions from the ML algorithms, which showed that all of the important variables used for classification modes had significant differences between the observation and control groups. First, a normality test was carried out, and the results showed that all the P values of the five indicators were less than 0.001 (data not shown), and none of them had passed the normality test, so they did not follow the normal distribution. Therefore, a nonparametric test should be used for further tests. The P values of the indicators were all less than 0.001, expect for PLT (Table 4), which indicated that the distributions of the two groups on the important indicators were different. The results proved the reliability of the classification model built by ML algorithms.
Full table
Discussion
ML is classified into three paradigms based on the targets: supervised, unsupervised, and reinforcement learning (32). ML algorithms display improved predictive function and low error in certain study fields, allowing the extraction of clinically relevant information from test data (33-36). In particular, ML classifiers have already demonstrated strong performance in image-based diagnoses. However, the analysis of diverse and test data remains challenging. Here, we show that ML classifiers tackle test data in a manner similar to the hypothetic-deductive reasoning used by tests and unearth associations that previous statistical methods have not found.
In the study, by applying ML approaches, we have developed several ML-based prediction models for postoperative blood coagulation of children with CHD. We chose the algorithms with classical methods and excellent application practices to design an analytical method. As previously, Decision tree is a regular and useful classification method in ML based on a global optimal solution. Naive Bayes is an analytical method based on conditional probability, which is effective in predicting most datasets (37). SVM is an excellent technology with independent integrity theory (38). SVM, Random Forest and AdaBoost are “black box” models with response is opaque to us. Among them, the AUCs ranged from 0.81 to 0.84 in this study. These results indicate that the ML-driven coagulation model is more abundant and more robust than the model developed using traditional statistical methods.
From a classical and methodological perspective, we provide proof of concept for clinicians, especially cardiologists (CHD), in adopting the use of ML predictive models. With digitalization of electronic medical records, especially test data, and higher accessibility to biologic and genetic patient data, the data we have on patients is exponentially growing. Therefore, it will be necessary for techniques similar to the ones described. From a clinical perspective, candidate risk factors for postoperative abnormal blood coagulation were evaluated and ranked. Some factors, such as MCV, MCH, and MCHC, are modifiable and could serve as targets for therapeutic intervention by blood transfusion, such as plasma or platelet transfusion.
In a wide range of medical diagnostic models, age and sex are the two most basic variables, which are related to most diagnostic predictions and are even closely related. Controlled studies based on a large number of retrospective clinical data from the real world are different from prospective studies in the controllability of all variables involved in our study. The differences between the observation group and the control group in age and sex are general and popular, which has resulted in great difficulties in comparative studies.
All cases of postoperative abnormal blood coagulation with CHD children screened out from 1,690 samples were selected as the observation group. According to the distribution characteristics of sex and age in the control group, CHD children who had normal coagulation and similar distribution characteristics of sex and age were randomly selected as the control group from test data in order to make the statistical analysis of the two groups more effective and reliable. However, the age and sex still play important roles in feature exploration. The age and sex will be included in the subsequent classification model for classification prediction performance.
We further explored a new combination of indicators that could distinguish between normal and abnormal blood coagulation. A series of baseline classification models for distinguishing between postoperative normal blood coagulation and abnormal blood coagulation in children with CHD were established. Through continuous improvement and adjustment, optimal prediction models for abnormal blood coagulation in children with CHD were finally obtained. Data mining and statistical algorithms were used to evaluate the effect of the models. The best predictive model for abnormal coagulation in CHD children was built based on test data. By analyzing 1,690 cases, we found that the classification models built with variables including age and sex had an average classification accuracy of 75.79%. The AUCs for those models ranged from 0.81 to 0.83.
In addition, to further explore the better models, the Decision tree-based algorithm was used to explore the important features related to postoperative blood coagulation of children with CHD. WBC and PLT were further selected as important predictors and used the seven prioritized features for modeling on the basis of data-driven and medical selection, the same as primary objective prediction (Table 3). Among them, the TPR (sensitivity) of the AdaBoost, Random Forest and SVM prediction models reached more than 80% in the ROC curve. The AUCs of three models above also indicated good performance. With the combination of calibration curves and Lift curves, the better fit is the SVM model, which predicted postoperative abnormal coagulation and postoperative normal coagulation more accurately than the simple prediction. Further, the difficulty of screening for abnormal coagulation in CHD children is reduced, that is, the general test indicators can achieve a relatively accurate screening result of abnormal coagulation in CHD children, further reducing the cost and difficulty of screening for abnormal coagulation in CHD children.
The process of data analysis in this study included variable exploration, variable verification, model establishment and model verification. By applying data mining algorithms such as SVM, we obtained an excellent classification model. The reliability could then be proven by data mining algorithms and statistical tests. These may provide a new angle of thought for clinical scientific research. In addition to those quantitative indicators mentioned above, which mainly refer to numeric biochemical indicators obtained using data mining algorithms, there are also many other available data types, such as text analysis for imaging reports or other analyses for medical images, and so on. With increasing data dimensions, a further increase in model accuracy is expected.
Conclusions
The coagulation status of children with CHD may change after operation, so it is very important to predict coagulation abnormality early. The SVM coagulation model exhibited an improved predictive performance (the highest sensitivity) for the postoperative blood coagulation state of children with CHD, and the age, sex, MCV, MCH, MCHC, WBC and PLT may be the key features for prediction. Further prospective multicenter studies with multiple datasets are needed to confirm our results and to reduce the influence of the imbalance in the target variables. Moreover, predicting blood product transfusion requirements during perioperative period of CHD children remains difficult. Further research may help determine the role of cardiac function in the treatment strategy of coagulation function as guidance, such as plasma or platelet transfusion. Based on the application of ML technology and data mining algorithms, combined with the large quantities of clinical data in the real world, using relevant medical knowledge to carry out independent mining exploration processes can provide a new reference basis for the early diagnosis of coagulation function and a new perspective for medical research.
Acknowledgments
We acknowledge colleagues from F&E Data Technology (Tianjin) Corporation for providing technical support in machine learning model building. We thank Xiaohuan Wang, Lijuan Qiu, Hua Shao, Qian Liu, Yu Liu, Shuaihang Zhang and Zijian Niu from the Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health, Beijing, China for all their invaluable efforts.
Funding: This work was supported by grants from the Cultivation Fund of Capital Medical University (PYZ19033) and the Cultivation Fund Project of the National Natural Science Foundation in Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health of China (GPY201802). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the TRIPOD Checklist: Prediction Model Development. Available at http://dx.doi.org/10.21037/tp-20-238
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-238
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-238). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Beijing Children’s Hospital, Capital Medical University gave expedited approval to review and use the medical data, such as electronic health records (ID: 2018-126). The pediatric patients or their guardians were not required to provide written informed consent for our study because of the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Williams LM, Hack LM. A precision medicine-based, 'fast-fail' approach for psychiatry. Nat Med 2020;26:653-4. [Crossref] [PubMed]
- Wang M, Zhou Y, Zong Z, et al. A precision medicine approach to managing 2019 novel coronavirus pneumonia. Precis Clin Med 2020;3:14-21. [Crossref] [PubMed]
- Mills S. Electronic health records and use of clinical decision support. Crit Care Nurs Clin North Am 2019;31:125-31. [Crossref] [PubMed]
- He J, Baxter SL, Xu J, et al. The practical implementation of artificial intelligence technologies in medicine. Nat Med 2019;25:30-6. [Crossref] [PubMed]
- Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25:24-9. [Crossref] [PubMed]
- Van Calster B, Wynants L. Machine learning in medicine. N Engl J Med 2019;380:2588. [Crossref] [PubMed]
- Deo RC. Machine learning in medicine. Circulation 2015;132:1920-30. [Crossref] [PubMed]
- Bizopoulos P, Koutsouris D. Deep learning in cardiology. IEEE Rev Biomed Eng 2019;12:168-93. [Crossref] [PubMed]
- Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol 2019;16:391-403. [Crossref] [PubMed]
- Wong NC, Shayegan B. Patient centered care for prostate cancer-how can artificial intelligence and machine learning help make the right decision for the right patient? Ann Transl Med 2019;7:S1. [Crossref] [PubMed]
- Trojano M, Tintore M, Montalban X, et al. Treatment decisions in multiple sclerosis - insights from real-world observational studies. Nat Rev Neurol 2017;13:105-18. [Crossref] [PubMed]
- Marx V. Biology: The big challenges of big data. Nature 2013;498:255-60. [Crossref] [PubMed]
- Bender E. Big data in biomedicine: 4 big questions. Nature 2015;527:S19. [Crossref] [PubMed]
- Frieden TR. Evidence for health decision making - beyond randomized, controlled trials. N Engl J Med 2017;377:465-75. [Crossref] [PubMed]
- Bates DW, Saria S, Ohno-Machado L, et al. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood) 2014;33:1123-31. [Crossref] [PubMed]
- Razavian N, Blecker S, Schmidt AM, et al. Population-Level Prediction of Type 2 Diabetes From Claims Data and Analysis of Risk Factors. Big Data 2015;3:277-87. [Crossref] [PubMed]
- Medrano López C, Guia Torrent JM, Rueda Nunez F, et al. Update on pediatric cardiology and congenital heart disease. Rev Esp Cardiol 2009;62 Suppl 1:39-52. [PubMed]
- Williams K, Carson J, Lo C. Genetics of Congenital Heart Disease. Biomolecules 2019;9:879. [Crossref] [PubMed]
- Houdijk WP, Van Den Besselaar AM. International multicenter international sensitivity index (ISI) calibration of a new human tissue factor thromboplastin reagent derived from cultured human cells. J Thromb Haemost 2004;2:266-70. [Crossref] [PubMed]
- Rowe M. An introduction to machine learning for clinicians. Acad Med 2019;94:1433-6. [Crossref] [PubMed]
- Thamaraiselvi G, Kaliammal A. Data mining: Concepts and techniques. SRELS Journal of Information Management 2004;41:339-48.
- Che D, Liu Q, Rasheed K, et al. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv Exp Med Biol 2011;696:191-9. [Crossref] [PubMed]
- Heikamp K, Bajorath J. Support vector machines for drug discovery. Expert Opin Drug Discov 2014;9:93-104. [Crossref] [PubMed]
- Taha AM, Mustapha A, Chen SD. Naive bayes-guided bat algorithm for feature selection. ScientificWorldJournal 2013;2013:325973. [Crossref] [PubMed]
- Breiman L. Random forests. Machine Learning 2001;45:5-32. [Crossref]
- Bjurgert J, Valenzuela PE, Rojas CR. On adaptive boosting for system identification. IEEE Trans Neural Netw Learn Syst 2018;29:4510-4. [Crossref] [PubMed]
- Shouval R, Hadanny A, Shlomo N, et al. Machine learning for prediction of 30-day mortality after ST elevation myocardial infraction: An acute coronary syndrome israeli survey data mining study. Int J Cardiol 2017;246:7-13. [Crossref] [PubMed]
- Chiew CJ, Liu N, Wong TH, et al. Utilizing machine learning methods for preoperative prediction of postsurgical mortality and intensive care unit admission. Ann Surg 2020;272:1133-9. [Crossref] [PubMed]
- Luo Y, Tang Z, Hu X, et al. Machine learning for the prediction of severe pneumonia during posttransplant hospitalization in recipients of a deceased-donor kidney transplant. Ann Transl Med 2020;8:82. [Crossref] [PubMed]
- Darst BF, Malecki KC, Engelman CD. Using recursive feature elimination in random forest to account for correlated variables in high dimensional data. BMC Genet 2018;19:65. [Crossref] [PubMed]
- Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: A multidisciplinary view. J Med Internet Res 2016;18:e323. [Crossref] [PubMed]
- Baştanlar Y, Ozuysal M. Introduction to machine learning. Methods Mol Biol 2014;1107:105-28. [Crossref] [PubMed]
- Senders JT, Staples PC, Karhade AV, et al. Machine Learning and Neurosurgical Outcome Prediction: A Systematic Review. World Neurosurg 2018;109:476-486.e1. [Crossref] [PubMed]
- Heo J, Yoon JG, Park H, et al. Machine learning-based model for prediction of outcomes in acute stroke. Stroke 2019;50:1263-5. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Meyer A, Zverinski D, Pfahringer B, et al. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir Med 2018;6:905-14. [Crossref] [PubMed]
- Zhang Z. Naive bayes classification in R. Ann Transl Med 2016;4:241. [Crossref] [PubMed]
- Huang S, Cai N, Pacheco PP, et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 2018;15:41-51. [PubMed]


