
A novel missense mutation in SMPX causes a rare form of X-linked postlingual sensorineural hearing loss in a Chinese family
Introduction
Hearing loss has always been a worldwide public health problem. It is estimated that 466 million people (6.1% of the world’s population) suffer from hearing loss, and unaddressed hearing loss costs the global economy US$750 billion annually (WHO data, www.who.int). Major causes of hearing loss include genetic etiological agents, chronic middle ear infections, noise-induced hearing loss, and ototoxic drugs. Approximately 60% of hearing loss cases are hereditary hearing loss, which can be divided into syndromic hearing loss (SHL) and non-syndromic hearing loss (NSHL) (1). The X-linked forms are far rarer than autosomal or mitochondrial-inherited hearing loss, accounting for only 1–5% of all reported mutations associated with NSHL (2). Till now, in X-linked NSHL, only six loci (DFNX1–6) and five genes have been reported in the literature (3).
SMPX (DFNX4, OMIM 300066), mapping to Xp.22.1, is composed of five exons and encodes a small muscle protein comprising 88 amino acids, which is originally cloned from skeletal muscle without known functional domains (4,5). Initial studies have shown that this gene is highly expressed in mammalian fetal and adult skeletal muscles and hearts, suggesting that it acts as a mechanical transducer during sensory changes in mechanical loads (6). However, neither Smpx gene knockout nor overexpression animal models have significant muscle dysfunction phenotypes (7). In the following study, it is confirmed that SMPX mutations are associated with X-linked hearing loss with obvious clinical and genetic heterogeneity (8-16). Moreover, SMPX has been identified in fetal and adult inner ears (9). Yoon et al. (17) reported the expression of mouse Smpx in the inner ears and vestibular hair cells. Ghilardi et al. (18) speculated that smpx localizes to the cuticular plate of the inner ear hair cells in zebrafish. On the other hand, in vitro studies suggested small muscle protein is mainly involved in regulation of cytoskeletal dynamics and promotion of myocyte fusion through the Rac1-p38 and IGF-1 pathways (6,7). Ferrán et al. (19) reported that the nuclear receptor NOR-1 positively regulates small muscle protein and myotube differentiation. Considering the expression pattern of SMPX and its response to mechanical force, it is believed that SMPX may play an important role in protecting the stereocilia of the sensory epithelium from mechanical stress during hearing (8).
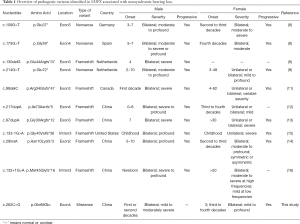
To date, three nonsense mutations and seven frameshift mutations in SMPX have been reported to be associated with X-linked NSHL (Table 1). However, the missense mutation of SMPX has never been reported before. We investigated X-linked hearing loss in a four-generation Chinese family and identified a novel missense mutation (c.262C>G: p.Gln88Glu) in SMPX by whole-exome sequencing (WES). Also, we found the proband carries two new compound heterozygous mutations (c.9259G>A: p.Val3087Ile and c.8576G>A: p.Arg2859His) in the USH2A gene. Furthermore, skewed X-chromosome inactivation may be one of the reasons for phenotypic heterogeneity.
Full table
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tp-20-435).
Methods
Subjects and clinical assessment
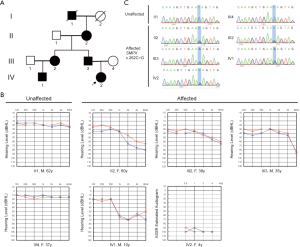
This study investigated X-linked hearing loss in a four-generation Chinese family with a total of 10 people, including five patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This research was approved by the Institutional Ethics Committee of The Second Hospital of Jilin University. The study was conducted after receiving written informed consent from the patients. The proband (IV-2, Figure 1A), a 4-year-old female, was diagnosed with profound sensorineural hearing loss at the Department of Otolaryngology, the Second Hospital of Jilin University. The hearing loss began when she was approximately 3 years old, and it was more severe than that of other members of the family. A detailed clinical evaluation of all participating family members was obtained and collected by a questionnaire, which included age of onset, noise exposure, subjective degree of hearing loss, progression, treatment, and other relevant clinical manifestations. Physical and otoscopic examinations, acoustic immittance testing, pure tone audiometry (PTA), and vestibular testing were used to assess the function of the middle ear and vestibular system as well as hearing sensitivity. The proband underwent auditory brainstem response (ABR), multiple frequency auditory steady-state evoked responses (ASSR), distortion product otoacoustic emission (DPOAE), high-resolution computed tomography (HRCT), magnetic resonance imaging (MRI), ophthalmic examination, and routine blood examination. The degree of hearing loss (normal, mild, moderate, moderately severe, severe, and profound) was classified and defined according to the recommendations of the Global Burden of Disease Expert Group on Hearing Loss (20).
Deafness gene hotspot mutation screening
A mutation analysis of the proband was performed on 20 common variants of four genes related to hearing loss: GJB2, GJB3, SLC26A4, and mtDNA (GJB2 c.35delG, c.167delT, c.176_191del16GCTGCAAGAACGTGTG, c.235delC, c.299_300delAT; GJB3 c.538C>T, c.547G>A; SLC26A4 c.281C>T, c.589G>A, C.919-2A>G, c.1174A>T, c.1226G>A, c.1229C>T, c.1707+5G>A, c.1975G>C, c.2027T>A, c.2162C>T, c.2168A>G, and 12S rRNA A1555G, C1494T).
WES and bioinformatic analysis
Genomic DNA was extracted from the peripheral blood sample of all participants through the standard phenol/chloroform method. WES was performed on two affected subjects (III-3 and IV-2) and one normal subject (III-4) by the Beijing Genomics Institution, China. The DNA sample was randomly broken into fragments with an average DNA size of 180–280 bp. Each targeted exon was sequenced using an Illumina HiSeq2000 platform after enrichment, hybridization, and capture. Mapping and analysis were based on the human reference genome (UCSC hg19). The variants were filtered and identified by using public databases, including dbSNP, 1000 Genomes Project, ESP6500, ExAC, and the human gene mutation database (HGMD). Then, the synonymous variants and irrelevant gene variants that did not match clinical manifestations and inheritance patterns were further excluded to narrow down the number of the candidate variants. The functional prediction was carried out using PROVEAN, PolyPhen-2, Mutation Taster, and I-Mutant 3.0.
Sanger sequencing
The SMPX (Gene ID: 23676) variant was amplified and sequenced to confirm if the gene co-separated with the hearing loss phenotype in the family (Forward primers: 5'-CAATGGATCTATGACTG-3'; Reverse primers: 5'-CCAAAGAGATCAAATGT-3'). Sanger sequencing was performed on the ABI 3730xl DNA Analyzer.
X-chromosome inactivation assay
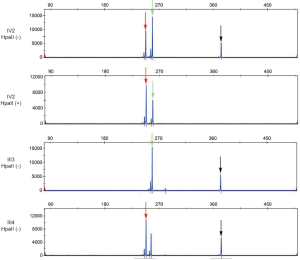
X-chromosome inactivation analysis was performed on the proband (IV-2) and her parents (III-3 and III-4). An evaluation of X-chromosome inactivation based on HpaII methylation analysis, which is a polymorphic short tandem repeat (STR) in the X-linked AR gene of peripheral blood leukocytes (21) was performed. The peripheral blood DNA sample was digested with HpaII (R0171S, NEB) at 37 °C overnight, and then at 95 °C for 10 min for enzyme inactivation. For each sample, 200 ng of DNA was used for polymerase chain reaction (PCR) with specific primers of the methylation regions of the AR gene. The fluorescently labelled PCR products were analyzed by capillary gel electrophoresis to determine the Xi level of each allele by calculating the ratio between the heights of two peaks.
Results
Clinical characteristics
The proband (Figure 1A, IV-2) who passed the newborn hearing screening test (NHST) began to experience hearing loss in her 3rd year. A detailed audiological test and imaging examination were performed when she was 4 years old. Neither ear passed DPOAE. An ABR test showed that the V wave response of both ears was normal, and the V wave response thresholds were 95 dB nHL (left) and 90 dB nHL (right). An ASSR test also showed that she exhibited a bilateral profound sensorineural hearing loss (Figure 1B), though HRCT as well as MRI showed no abnormalities. Ophthalmic examination and routine blood examination were performed at the same time, but no abnormal symptoms were found.
The audiological-related characteristics of all participants in the family, assessed by subjective and objective audiological examinations and summarized in Table 2, showed significant heterogeneity. While PTA data revealed that all affected family members exhibited binaural symmetrical sensorineural hearing impairment, and that high-frequency hearing loss occurred more severely than low-frequency hearing loss, the degree of hearing loss and the age of onset were quite different between male and female. Male patients suffered from hearing loss in the first to second decades, about 10 to 20 years earlier than female patients except the proband. The father (III-3) and cousin (IV-1) of the proband exhibited mild to moderately severe hearing loss, which was more severe than in female patients (II-2, III-2) of the same age. Subject II-2 had intermittent tinnitus, often accompanied by further hearing loss. None of the patients in the family had a history of vestibular dysfunction, noise exposure, or ototoxic medication use.

Full table
Mutation analysis
We obtained 5.08 Gb of raw data. The coverage of the targeted bases was 99.16%, and approximately 96.34% of the targeted region was covered at least 30×. After filtering and analyzing the data according to the method described in the materials and methods, 32 variants remained, of which 16 variants followed autosomal dominant inheritance pattern and one followed X-linked dominant inheritance pattern. We then eliminated autosomal dominant genes including DIAPH1, DIAPH3, DSPP, GRHL2 based on the family clinical phenotype. The variant in SMPX (c.262C>G: pGln88Glu) was finally identified as possibly related to the post-lingual hearing loss of the family. Furthermore, this missense mutation has not been reported before and was absent in 300 normal controls. Sanger sequencing validated that it was found in all of the affected members (II-2, III-2, III-3, IV-1 and IV-2), but absent in unaffected members (Figure 1C). In other words, c.262C>G located in exon 4 of SMPX was fully co-segregated with the hearing loss phenotype of this family. Also, we found that the proband carries two compound heterozygous mutations (c.9259G>A: p.Val3087Ile and c.8576G>A: p.Arg2859His) in the USH2A gene, but without any other symptoms except profound sensorineural hearing loss to date.
Table 3 shows the prediction results of the pathogenicity of these genetic alterations by online bioinformatics prediction tools (PolyPhen-2, Mutation Taster, and I-Mutant 3.0). The 88 amino acids in human SMPX are conserved across several species (Figure 2).

Full table

Analysis of X-chromosome inactivation skewing
Xi = a:b, where a represented the ratio of the height of the parent peaks of HpaII-digested to HpaII-undigested, similarly, b denoted the ratio of the height of the two maternal peaks. The X-inactivation ratios of paternal (a) to maternal (b) allele of the profound was 29:71, which means that approximately 71% of the paternal X-chromosome including the defective SMPX gene was active in the proband (Figure 3). Briefly, it indicated moderate skewing in the proband, which may account for the phenotypic differences.
Discussion
X-linked non-syndromic hearing loss is highly heterogeneous in genetic and clinical phenotypes. DFNX4 (DFN6) was first reported in 1996 in a Spanish family with X-linked dominant progressive non-syndromic sensorineural deafness (22). High-frequency hearing loss occurred in the affected males at school age and gradually developed into full-frequency severe-to-profound deafness before adulthood, while the carrier females showed moderate high-frequency deafness in their fourth decade. In 2011, Huebner et al. (8) identified two nonsense mutations (SMPX c.109G>T: p.Glu37*; SMPX c.175G>T: p.Gly59*) in German and Spanish families, respectively. Males presented with progressive, moderate or above hearing loss and onset at the age of 3–7 years. The age of onset in female carriers ranged from the second to the third decade, and hearing loss progressed to severe within 10–15 years. In the same year, Schraders et al. (9) reported a nonsense mutation (SMPX c.214G>T: p.Glu72*) and a frameshift mutation (SMPX c.130delG: p.Glu44Argfs*37) in two Dutch families. The age of onset for male and female carriers was 2–10 years (average, 3.3 years) and 3–48 years (average, 28.2 years), respectively. The men showed a severe increase in threshold values mainly during the first two decades, whereas women demonstrated significant inter-individual heterogeneity in age of onset, disease progression, degree of hearing loss, and inter-aural asymmetry in thresholds. In 2013, Abdelfatah et al. (11) confirmed a frameshift mutation (SMPX c.99delC: p.Arg34Glufs*47) in two multiplex families from Newfoundland. Hearing loss in males appeared to be similar to previous studies in degree and age of onset, but congenital hearing loss was not definitely excluded due to a non-detectable mild hearing loss during newborn screening. Likewise, female carriers displayed highly variable phenotypes, including a widespread age of onset (4–62 years), variable severity, and unilateral or bilateral symmetric or asymmetric deafness. In 2017, Niu et al. (12) first reported a novel frameshift mutation (SMPX c.217dupA: p.Ile73Asnfs*5) in a Chinese population with late-onset progressive NSHL, as well as congenital hearing impairment which had never been proven before. Subsequently, a North American family with a novel frameshift mutation creating a premature stop codon (c.133-1G>A: p.Gly45fs*36) of SMPX was described by Niu et al. (15). Meanwhile, Deng et al. (13), Gao et al. (14) and Lv et al. (16) successively identified three frameshift mutations of SMPX (c.87dupA: p.Gly30Argfs*12; c.29insA: p.Asn10Lysfs*3; c.132+1G>A: p.Met45Glyfs*16) from multiple Chinese lineages. To sum up, most pathogenic variants of SMPX caused post-lingual progressive NSHL, and a few led to congenital deafness. The affected males showed more severe hearing loss at a younger age, while female carriers manifested incomplete penetrance and variable expressivity probably due to skewed X-chromosome inactivation.
Generally, one of the two X chromosomes of female cells is randomly inactivated in mammals, resulting in most normal females being chimeras, that is, approximately half of the cells express the alleles on the paternal X chromosome, and the other half express the maternal. Non-random inactivation of the X chromosome, namely skewed X-chromosome inactivation, occurs when the ratio of paternal and maternal cell inactivation is highly deviated from 1:1, which might lead to the phenotypic heterogeneity of female carriers of X-linked inherited deafness. The human androgen receptor assay (HUMARA), based on HpaII methylation analysis of a polymorphic short tandem repeat (STR) in the X linked AR (androgen receptor) gene, is one of the most convenient, rapid and widely accepted methods for measuring X-chromosome inactivation (23).
In this study, we reported X-linked hearing loss in a four-generation Chinese family and identified a novel missense mutation (c.262C>G, p.Gln88Glu) in SMPX by WES. This variant was co-segregated with the post-lingual hearing loss phenotype and was absent in 300 normal controls. However, the female proband presented profound sensorineural deafness with early age of onset, which was significantly heterogeneous from other female carriers in the family. Analysis of the X-chromosome inactivation indicated moderate skewing in the proband, which was probably related to the heterogeneity of clinical characteristics.
In addition, we found the proband carries two new compound heterozygous mutations (c.9259G>A: p.Val3087Ile and c.8576G>A: p.Arg2859His) in the USH2A gene, but to date without any other symptoms except profound sensorineural hearing loss. USH2A (OMIM 608400), together with ADGRV1 and WHRN, is known to be associated with Usher syndrome type II (USH2). USH2A, located on chromosome 1q41, encodes the usherin which is primarily expressed on the basement membrane of the cochlea and retina and is involved in postnatal development of cochlear hair cells and retinal photoreceptors (24). USH2A variants are associated with autosomal recessive syndromic hearing loss with moderate to severe bilateral sensorineural hearing impairment and progressive visual loss that occurs during adolescence or beyond. So far, more than 600 mutations have been found in the USH2A gene, including nonsense mutations, frameshift mutations, splice-site mutations, missense mutations, and intron mutations (http://www.lovd.nl/USH2A). The mutation (USH2A c.9259G>A: p.Val3087Ile) has been reported in patients (25,26), while the variant (USH2A c.8576G>A: p.Arg2859His) has never been detected before. Whether the combination of USH2A and SMPX mutations causes profound hearing loss in the proband still needs long-term follow-up and further experimental research.
Conclusions
We reported X-linked hearing loss in a four-generation Chinese family and identified a novel missense mutation (c.262C>G, p.Gln88Glu) in SMPX by WES. This is the first study to report a missense mutation of SMPX in a Chinese family. Our findings have enriched the mutation and phenotypic spectrum of the SMPX gene. Furthermore, skewed X-chromosome inactivation may be one of the reasons for phenotypic heterogeneity.
Acknowledgments
We sincerely thank the family members who participated in this study.
Funding: This study has been funded by the Natural Science Foundation of Jilin Province Science and Technology Commission (20190201040JC).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tp-20-435
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-435
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-435). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This research was approved by the Institutional Ethics Committee of The Second Hospital of Jilin University. The study was conducted after receiving written informed consent from the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med 2006;354:2151-64. [Crossref] [PubMed]
- Petersen MB, Wang Q, Willems PJ. Sex-linked deafness. Clin Genet 2008;73:14-23. [Crossref] [PubMed]
- Corvino V, Apisa P, Malesci R, et al. X-Linked Sensorineural Hearing Loss: A Literature Review. Curr Genomics 2018;19:327-38. [Crossref] [PubMed]
- Patzak D, Zhuchenko O, Lee CC, et al. Identification, mapping, and genomic structure of a novel X-chromosomal human gene (SMPX) encoding a small muscular protein. Human Genetics 1999;105:506-12. [Crossref] [PubMed]
- Kemp TJ, Sadusky TJ, Simon M, et al. Identification of a novel stretch-responsive skeletal muscle gene (Smpx). Genomics 2001;72:260-71. [Crossref] [PubMed]
- Schindeler A, Lavulo L, Harvey RP. Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp Cell Res 2005;307:367-80. [Crossref] [PubMed]
- Palmer S, Groves N, Schindeler A, et al. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J Cell Biol 2001;153:985-98. [Crossref] [PubMed]
- Huebner AK, Gandia M, Frommolt P, et al. Nonsense mutations in SMPX, encoding a protein responsive to physical force, result in X-chromosomal hearing loss. Am J Hum Genet 2011;88:621-7. [Crossref] [PubMed]
- Schraders M, Haas SA, Weegerink NJ, et al. Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am J Hum Genet 2011;88:628-634. [Crossref] [PubMed]
- Weegerink NJ, Huygen PL, Schraders M, et al. Variable degrees of hearing impairment in a Dutch DFNX4 (DFN6) family. Hear Res 2011;282:167-77. [Crossref] [PubMed]
- Abdelfatah N, Merner N, Houston J, et al. A novel deletion in SMPX causes a rare form of X-linked progressive hearing loss in two families due to a founder effect. Hum Mutat 2013;34:66-9. [Crossref] [PubMed]
- Niu Z, Feng Y, Mei L, et al. A novel frameshift mutation of SMPX causes a rare form of X-linked nonsyndromic hearing loss in a Chinese family. PLoS One 2017;12:e0178384. [Crossref] [PubMed]
- Deng Y, Niu Z, Fan L, et al. A novel mutation in the SMPX gene associated with X-linked nonsyndromic sensorineural hearing loss in a Chinese family. J Hum Genet 2018;63:723-30. [Crossref] [PubMed]
- Gao S, Jiang Y, Wang G, et al. Skewed X-chromosome inactivation and next-generation sequencing to identify a novel SMPX variants associated with X-linked hearing loss in a Chinese family. Int J Pediatr Otorhinolaryngol 2018;113:88-93. [Crossref] [PubMed]
- Niu Z, Yan D, Bressler S, et al. A novel splicing mutation in SMPX is linked to nonsyndromic progressive hearing loss. Int J Pediatr Otorhinolaryngol 2018;104:47-50. [Crossref] [PubMed]
- Lv Y, Gu J, Qiu H, et al. Whole-exome sequencing identifies a donor splice-site variant in SMPX that causes rare X-linked congenital deafness. Mol Genet Genomic Med 2019;7:e967. [Crossref] [PubMed]
- Yoon H, Lee DJ, Kim MH, et al. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol 2011;44:69-78. [Crossref] [PubMed]
- Ghilardi A, Diana A, Prosperi L, et al. Expression pattern of the small muscle protein, X-linked (smpx) gene during zebrafish embryonic and larval developmental stages. Gene Expr Patterns 2020;36:119110. [Crossref] [PubMed]
- Ferrán B, Marti-Pamies I, Alonso J, et al. The nuclear receptor NOR-1 regulates the small muscle protein, X-linked (SMPX) and myotube differentiation. Sci Rep 2016;6:25944. [Crossref] [PubMed]
- Stevens G, Flaxman S, Brunskill E, et al. Global and regional hearing impairment prevalence: an analysis of 42 studies in 29 countries. Eur J Public Health 2013;23:146-52. [Crossref] [PubMed]
- Plenge RM, Hendrich BD, Schwartz C, et al. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet 1997;17:353-356. [Crossref] [PubMed]
- del Castillo I, Villamar M, Sarduy M, et al. A novel locus for non-syndromic sensorineural deafness (DFN6) maps to chromosome Xp22. Hum Mol Genet 1996;5:1383-7. [Crossref] [PubMed]
- Busque L, Zhu J, DeHart D, et al. An expression based clonality assay at the human androgen receptor locus (HUMARA) on chromosome X. Nucleic Acids Res 1994;22:697-8. [Crossref] [PubMed]
- Liu X, Bulgakov OV, Darrow KN, et al. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc Natl Acad Sci U S A 2007;104:4413-8. [Crossref] [PubMed]
- Miyagawa M, Naito T, Nishio SY, et al. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS One 2013;8:e71381. [Crossref] [PubMed]
- Huang L, Mao Y, Yang J, et al. Mutation screening of the USH2A gene in retinitis pigmentosa and USHER patients in a Han Chinese population. Eye (Lond) 2018;32:1608-14. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


