
A long non-coding RNAs expression signature to improve prognostic prediction of Wilms tumor in children
IntroductionOther Section
Wilms tumor (WT), also known as nephroblastoma, is the most frequent malignancy of the kidney in children, constituting nearly 90% of childhood renal tumors (1). Despite the development and advancement of various treatment approaches, 15% of WT patients undergo metastasis or relapse, suggesting poor prognosis after initial treatment (2). Therefore, it is of great clinical benefit to explore prognostic factors in children with different WT stages, which could provide valuable guidance on formulating an accurate therapeutic regime for each patient. Previous studies that were committed to identifying potential prognostic markers for WT patients relied on very few factors, mainly histology analysis and characterization of tumor stage, which has been deemed insufficient as WT in children was shown to be affected by a variety of factors, including genetic and epigenetic changes that underlie WT pathogenesis (1). The prognosis for WT children in clinical practice incorporates a diverse set of measurements, including age, tumor size, loss of heterozygosity (LOH) for chromosomes 16q and 1p, and sensitivity to medications (3-6). Moreover, radiation (7), surgery, microscopic residual disease, diffuse anaplasia (8), lymph node involvement (9), and a combination of lymph node and LOH status (10) have been associated with the prognosis of WT in children. Therefore, it is important to build a prognostic model using multiple variables that underlie a broad spectrum of genetic and clinicopathological factors.
Long non-coding RNAs (lncRNAs) are characterized as non-coding transcripts longer than 200 nucleotides (11). Next-generation and high-throughput sequencing techniques have enabled a significant breakthrough in lncRNA identification and characterization in recent years. Gene expression of various lncRNAs is dysregulated in numerous cancers, and it has been reported to correlate with cancer recurrence, metastasis, and poor prognosis (12-24). Previous studies have suggested that several lncRNAs, such as LINC00473 (25), MIAT (26), SOX21-AS1 (27), and LINP1 (28), play important roles in the pathogenesis of WT. However, it remains largely unknown whether lncRNAs could be used as prognostic markers of WT in children. The present study sought to characterize lncRNA signature as a robust prediction for WT children’s prognosis and construct a prognostic risk index based on comprehensive RNA-sequencing analysis. We established a prognosis model using RNA-sequencing data and subsequently developed and validated a genomic- clinicopathological nomogram that integrated risk scores and traditional clinicopathological factors. Moreover, Gene Set Enrichment Analysis (GSEA) revealed molecular pathways significantly enriched in different risk groups (29,30). Additionally, CIBERSORTx and ESTIMATE algorithms were applied to explore the immune microenvironment discrepancies between the high-risk and low-risk groups (31-33). Altogether, the current study presents a novel and efficient method for the prognosis of patients with WT and provides insight into the molecular mechanisms that affect WT’s prognosis. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tp-20-318).
MethodsOther Section
Study population and RNA-sequencing data processing
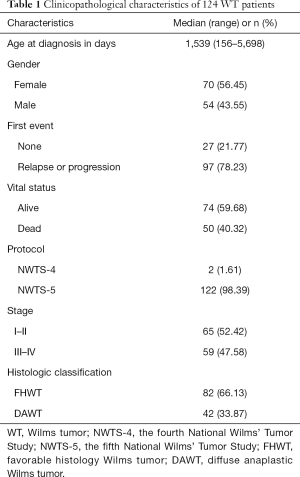
The level 3 RNAseq data of primary WT samples, including raw read counts and reads per kilobase million (RPKM), were obtained from the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA), and relevant clinical information of the WT patients was downloaded from the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) Data Matrix in April 2020. This study was conducted following the TARGET publication guidelines. The inclusion criteria of this study were as follows: (I) pathological diagnosis of primary WT; (II) clinical information including age at diagnosis, gender, cooperative group protocol, stage, histologic classification, the first event, event-free survival (EFS), vital status, and overall survival (OS) was documented; and (III) complete RNAseq data. The exclusion criteria were as follows: (I) patients with a history of other malignancies; (II) tumor tissues were not used for RNA sequencing analysis; (III) clinical information not available. Thus, 124 WT patients were selected and enrolled in this study (Tables S1,S2). Participant clinicopathological characteristics are listed in Table 1. The geneset annotation GENCODE Human (GRCh38.p13) was used to annotate protein-coding genes and lncRNAs were selected for further study (34). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Full table
Identification and validation of the prognostic lncRNA signature
The “survival” package in the R software (version 3.6.2; https://www.R-project.org/) was used to analyze RPKM data [transformed to log2(RPKM+1)] through univariate Cox regression analysis, and P<0.05 was used as the cut-off criterion. Next, through the “glmnet” package (version 3.0-2) in R, the least absolute shrinkage and selection operator (LASSO) Cox regression was performed to select key lncRNAs among the significant lncRNAs from univariate Cox analysis (35,36). We used 10 times cross-validations to determine the best penalty parameter lambda. The key lncRNAs selected by the LASSO method were considered as candidate variables for inclusion in the multivariate Cox analysis. Then, the independently prognostic lncRNAs were identified in the multivariate Cox analysis to obtain their coefficients (β values), with which the prognostic formula was constructed as follows: (β1 × log2(RPKM+1) value of gene1) + (β2 × log2(RPKM+1) value of gene2) + ... + (βn × log2(RPKM+1) value of genen). Subsequently, the risk score of each WT patient was calculated based on the prognostic formula. Then, the “surv_cutpoint” function in the “survminer” R package (version 0.4.6) was utilized to achieve the optimal cutoff values for risk scores. According to the cutoff values, we divided the 124 WT participants in the cohort into a low‐risk group and a high‐risk group. Between the low- and high-risk groups, differences in OS and EFS were examined by log-rank test, and Kaplan-Meier survival curves were constructed by the “survival” package in R software. A P value <0.05 was considered statistically significant.
Development and assessment of a predictive nomogram
Univariate and multivariate Cox regression analyses were conducted to ascertain whether the risk score of lncRNAs and the clinicopathological variables could be prognostic markers related to OS for WT patients, and a P<0.05 was deemed significant. Then, according to the results of Cox regression analyses, we constructed a genomic-clinicopathologic nomogram to predict 1‐, 3‐, and 5‐year OS through the “rms” package (version 5.1-4) in R software. Using the same R package, we generated a calibration curve to verify the consistency between the nomogram-predicted probability of OS and the actual OS. Furthermore, to assess the accuracy of the predictive nomogram, we performed time-dependent receiver operating characteristic (ROC) curve analyses and calculated the area under the curve (AUC) through the “timeROC” package (version 0.4) in R (37). Additionally, we conducted a decision curve analysis (DCA) to examine the clinical value with the “stdca.R” statistical code (38) in R.
Identification of differentially expressed genes and construction of protein-protein interaction network
Using the “DESeq2” package (version 1.26.0) in R, the differentially expressed genes, including protein-coding genes (DEPCGs) and lncRNAs (DELs) were identified by comparing the low-risk group and high-risk group of primary WT, with the screening conditions of |log2 fold change| >1, and a false discovery rate (FDR) or adjusted P-value <0.05. These DEPCGs were then used to construct the protein-protein interaction (PPI) network through the Search Tool for the Retrieval of Interacting Genes (STRING) online database (39). Required interaction score >0.4 was used as the cut-off criterion. Then, the data downloaded from STRING were used to model the PPI network through Cytoscape software (version 3.7.1, https://cytoscape.org) (40), and the densely connected regions were identified through the Molecular Complex Detection (MCODE) plug-in Cytoscape with the default criteria (degree cutoff =2, node score cutoff =0.2, Haircut = true, Fluff = false, K-score =2, and Max depth =100) (41).
GSEA
Before running GSEA, the normalization of the raw read counts of protein-coding genes was performed by “DESeq2”. Then, GSEA (version 4.0.3) was conducted to demonstrate significant differences in protein-coding genes between the low-risk and high-risk groups (29,30). Canonical pathway gene sets (c2.cp.v7.1.symbols.gmt) and Gene Ontology (GO) gene sets including biological process (BP) (c5.bp.v7.1.symbols.gmt), molecular function (MF) (c5.mf.v7.1.symbols.gmt), and cellular component (CC) (c5.cc.v7.1.symbols.gmt) were selected from the Molecular Signatures Database (MSigDB) as the reference gene sets (42-44). The number of permutations was set at 1,000, and the nominal P value <0.01 was considered statistically significant.
Analysis of immune infiltration level
The analytical tool CIBERSORTx provides an estimation of member cell types’ abundances in a mixed cell population, using gene expression data (31,32). We utilized CIBERSORTx to assess the proportions of 22 immune cell subtypes in the primary WT samples, using the RPKM data of protein-coding genes. The permutations for significance analysis were set at 1000. Then, we performed the 2-sided Wilcoxon test through the “ggpubr” package (version 0.2.5) in R, to compare differences in 22 immune cell subtypes between the low- and high-risk groups. Subsequently, through the “estimate” package (version 1.0.13), the immune and stromal scores were calculated based on the ESTIMATE algorithm (33), and Welch’s t-test performed the comparisons of the stromal and immune scores between the 2 groups with “ggstatsplot” package (version 0.4.0) in R. A P value < 0.05 denoted statistical significance.
Statistical analysis
The R software version 3.6.2 and several R packages were used for statistical analyses, and a 2-tailed P value <0.05 denoted statistical significance. The univariate and multivariate Cox regression analyses were performed by the “survival” package. The LASSO Cox regression analyses were calculated by the “glmnet” package (35,36), and 10 times cross-validations were used to determine the best penalty parameter lambda. Kaplan-Meier analyses and survival curves were constructed through the “survival” package. The “rms” package generated the nomogram and the calibration curve. The time-dependent ROC curve analyses were performed by the “timeROC” package (37).
ResultsOther Section
Identification and validation of the prognostic lncRNA signature
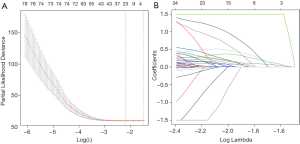
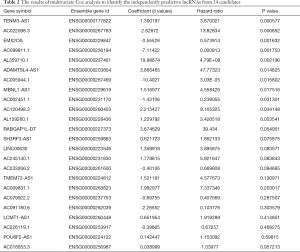
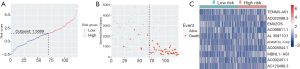
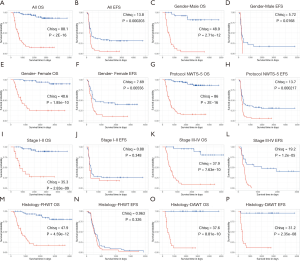
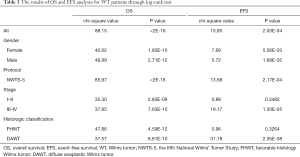
After filtering of low-expression lncRNAs, 2,927 lncRNAs were generated for further study. Subsequently, univariate Cox analysis was performed to obtain 244 lncRNAs associated with OS of WT patients (P<0.05). Among the 244 prognosis-associated lncRNAs, 24 were identified using the LASSO Cox method (Figure 1). Then, through multivariate Cox analysis, ten lncRNAs (TENM3-AS1, AC022098.3, EMX2OS, AC099811.1, AL359710.1, ADAMTSL4-AS1, AC005944.1, MBNL1-AS1, AC002451.1, and AC120498.3) were selected from 24 candidates of lncRNAs as independent predictors associated with OS (P<0.05), and the coefficients of the 10 lncRNAs were obtained (Table 2). The formula of the lncRNA-based prognostic index model was imputed as follows: [1.3×log2(RPKM+1) value of TENM3-AS1] + [2.627×log2(RPKM+1) value of AC022098.3] + [‒0.5553×log2(RPKM+1) value of EMX2OS] + [‒7.114 × log2(RPKM+1) value of AC099811.1] + [19.99×log2(RPKM+1) value of AL359710.1] + [3.866 × log2(RPKM+1) value of ADAMTSL4-AS1] + [‒10.4 × log2(RPKM+1) value of AC005944.1] + [1.517 × log2(RPKM+1) value of MBNL1-AS1] + [‒1.431×log2(RPKM+1) value of AC002451.1] + [2.215×log2(RPKM+1) value of AC120498.3]. Then, the risk score for each participant was calculated, and all participants were divided into high‐risk (n=54) and low‐risk (n=70) groups based on the optimal cutoff value (1.5069) for risk scores (Figure 2). Participants in the high‐risk group had a significantly worse OS and EFS than those in the low‐risk group (P<2E-16 and P=2.03E-04, respectively) (Figure 3A,B, Table 3). Importantly, subgroup analyses based on the clinicopathological variables (i.e., gender, cooperative group protocol, stage, and histologic classification) indicated that the prognostic value of the risk scores for OS and EFS was independent of all these clinicopathological variables, except for EFS in the subgroup of stage I-II and favorable histology Wilms tumor (FHWT) (P=0.348 and P=0.326; Figure 3C,D,E,F,G,H,I,J,K,L,M,N,O,P, Table 3).
Full table
Full table
Development and assessment of a predictive nomogram
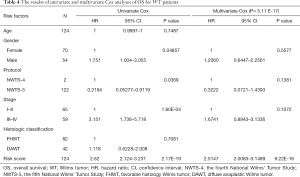
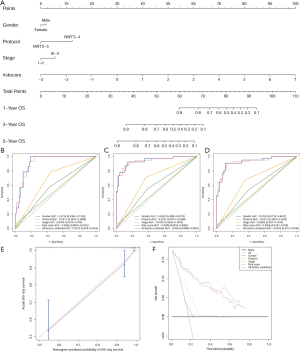
Based on the univariate Cox regression analysis results, the risk score of lncRNAs and 3 clinicopathological factors (i.e., gender, cooperative group protocol, and stage) were all significantly related to OS for WT patients (Table 4). The subsequent multivariate Cox analysis (P value for Wald test: 5.11E-17) further demonstrated that the risk score remained a powerful and independent prognostic factor (P=9.22E-16) (Table 4). Through integrating the 4 prognostic factors identified by univariate Cox analysis, a genomic-clinicopathologic nomogram was developed. This combined model is depicted in Figure 4A. The concordance index of the combined OS prediction model was 0.8780 (95% CI: 0.8407‒0.9154). The nomograms’ predictive accuracy was further evaluated by time-dependent ROC curves of 1‐, 3‐, and 5‐year OS among the models. The results suggested that the combined model and the risk score model had a significantly greater AUC than that of the gender, cooperative group protocol, and stage models (Figure 4B,C,D; Tables 5,6). Additionally, there was no significant difference in AUC between the combined model and the risk score model (1-year P=0.8610, 3-year P=0.7024, 5-year P=0.9354) (Table 6). Furthermore, the calibration curve also demonstrated high consistency between the actual proportion of 500-day OS and the nomogram-predicted probability of 500-day OS (Figure 4E). Meanwhile, DCA illustrated that the net benefit of the 500-day decision curve for the combined model was greater than that for the gender, cooperative group protocol, and stage model, further supporting the clinical value of the genomic-clinicopathologic nomogram (Figure 4F).
Full table
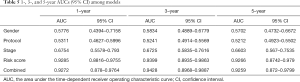
Full table
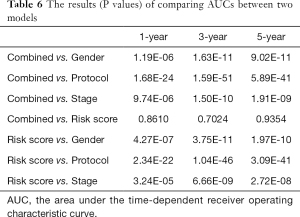
Full table
Identification of differentially expressed genes and construction of PPI network
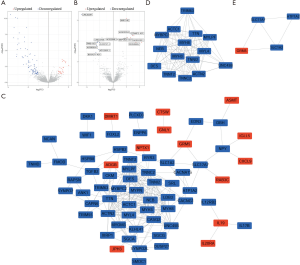
A total of 105 DEPCGs (24 upregulated and 81 downregulated) and 14 DELs (7 upregulated and 7 downregulated) were identified in the high-risk group (Figure 5A,B). Furthermore, the PPI network of the 105 DEPCGs was generated (Figure 5C). Following MCODE analysis, 2 densely connected regions in the PPI network were identified. The first region consisted of 15 target protein-coding genes, including MYL4, MYBPC2, ACTC1, MYLPF, DES, TRIM63, NEB, TTN, TNNI2, TNNC2, MYH3, UNC45B, ACTN2, MYH8, and TNNT2 (Figure 5D). The second region consisted of 4 target protein-coding genes, including ATP1A2, SLC1A2, SLC17A7, and GRM5 (Figure 5E).
GSEA
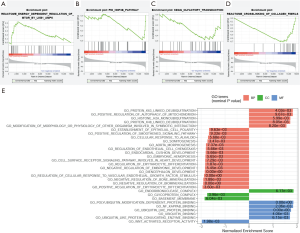
Through canonical pathway gene sets, GSEA identified energy-dependent regulation of mTOR by LKB1-AMPK from the Reactome subset, which was significantly enriched in the high-risk group [(nominal P=0.007859, normalized enrichment score (NES) = 1.728] (Figure 6A). By contrast, 3 other pathways, including the HNF3B pathway (nominal P=0.009434, NES = ‒1.631) from the PID subset (Figure 6B), olfactory transduction (nominal P=0.009634, NES = ‒1.599) from the Kyoto Encyclopedia of Gnese and Genomes (KEGG) subset (Figure 6C), and crosslinking of collagen fibrils (nominal P=0.009940, NES = ‒1.730) from the Reactome subset (Figure 6D), were significantly enriched in the low-risk group. Through GO gene sets, GSEA identified 11 GO terms consisted of 5 BP, 1 CC, and 5 MF, which were significantly enriched in the high-risk group (nominal P<0.01) (Figure 6E), and 19 GO terms consisted of 16 BP, 2 CC, and 1 MF, which were significantly enriched in the low-risk group (nominal P<0.01; Figure 6E).
Analysis of immune infiltration level
The CIBERSORTx analysis demonstrated that the proportion of monocytes activated dendritic cells and resting mast cells were significantly reduced in the high-risk group compared to that in the low-risk group (P=0.0464, P=0.0241, and P=0.00266), and the proportion of M1 macrophages (pro-inflammatory) in the low-risk group was substantially lower than that in the high-risk group (P=0.00944; Figure 7A). However, the stromal and immune scores showed no significance between groups (P=0.1339 and P=0. 8536; Figure 7B,C).

DiscussionOther Section
In clinical practice, it is apparent that tumor progression and prognosis of patients are affected by multiple factors, yet many studies have only focused on single or few clinicopathologic factors for WT patients’ prognosis (7-9). For instance, Tang et al. constructed nomograms to predict OS of children with WT based on 5 clinicopathologic factors, but AUCs of 3- and 5-year OS were not more than 0.74 (45). Nonetheless, prognostic tumor biomarkers also play a vital role in the diagnosis and treatment of tumors. For example, LOH for chromosomes 16q and 1p was demonstrated as a specific marker for increased relapse risk in favorable-histology WT (46). Several retrospective studies of heterogeneously-treated patients suggested an association between a gain of chromosomes 1q and tumor recurrence (47,48). Our previous study also indicated that astrocyte elevated gene-1 overexpression in histologically favorable WT was associated with poor patient prognosis (49). Also, Gong et al. have reported a 5-microRNA signature model to predict WT patients’ prognosis based on the TARGET database, but AUC was only 0.767 (50). Very few studies currently focus on the development of prognostic models that integrate the clinicopathologic features and biomarkers based on comprehensive sequencing analysis for WT patient prognosis.
In this study, we performed multiple analyses to identify the prognostic lncRNA signature, including filtering of low-expression lncRNAs, identification of lncRNAs associated with OS through univariate Cox analysis and LASSO method, and identification of independently predictive lncRNAs through multivariate Cox analysis. As a result, the potential minimum number of robust lncRNAs to predict WT patients’ prognosis was obtained. Accordingly, a risk-score formula was constructed, and the subsequent log-rank tests and Kaplan-Meier analyses further confirmed the predictive value of the risk score for OS and EFS in the low- and high-risk groups of WT patients. Among the 10 prognostic lncRNAs in the present study, the low expression of EMX2OS (also known as EMX2-AS1 or NCRNA00045) was correlated with a poor prognosis in participants with WT [β=‒0.55528, hazard ratio (HR) =0.573914; Table 2]. Interestingly, Gu et al. also reported that downregulation of EMX2OS was an independent prognosis factor for shorter recurrence-free survival of classical papillary thyroid cancer (51).
Nomograms also called nomographs or alignment charts, which are widely used as prognostic devices in the field of medicine and oncology, can generate the individual probability of clinical events by integrating multiple prognostic variables (52). In this study, a genomic- clinicopathologic predictive nomogram was developed through incorporating multiple prognostic factors including gender, cooperative group protocol, stage, and risk score to predict 1-, 3-, and 5-year OS of children with WT, which has shown stronger prediction with better AUCs (1-, 3-, and 5-year OS in the combined model of 0.9272, 0.9428, and 0.9259, respectively) (45,50). The combined model and the risk score model demonstrated significantly higher predictive accuracy, which was further verified by the time-dependent ROC curve and calibration curve analyses. Importantly, there was no significant difference in AUC between the combined and the risk score models, which indicated that the risk score played a critical role in the genomic-clinicopathologic nomogram. Hence, as a powerful and independent prognostic factor, the risk score achieved extremely high accuracy without combining clinicopathologic factors.
Nonetheless, several limitations need to be addressed in the present study. First, WT’s sample size was relatively small, which might lead to bias that could reduce the accuracy of the prognostic model. Second, an external validation cohort for the prognostic model was not available, and all the data analyzed were collected from the TARGET Data Matrix, which might also result in biased discovery. Moreover, the results of bioinformatics were not verified through reverse transcription polymerase chain reaction (RT-qPCR). Finally, besides mRNA sequencing data, other potential biomarkers such as LOH at 1p/16q, gain of 1q, microRNA, and DNA methylation were not integrated into the study.
ConclusionsOther Section
In summary, through constructing a genomic-clinicopathologic nomogram, we presented a novel and efficient method for the prognosis of patients with WT and provided insight into the molecular mechanisms that affect WT’s prognosis.
AcknowledgmentsOther Section
The results published here are based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (
Funding: This study was supported by Shandong Provincial Natural Science Foundation of China (Grant No. ZR2020MH048) and the Science and Technology Development Plan Project of Shandong Province, China (Grant No. 2019GSF108061).
FootnoteOther Section
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-318
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-318). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Treger TD, Chowdhury T, Pritchard-Jones K, et al. The genetic changes of Wilms tumour. Nat Rev Nephrol 2019;15:240-51. [Crossref] [PubMed]
- Brok J, Lopez-Yurda M, Tinteren H V, et al. Relapse of Wilms’ tumour and detection methods: a retrospective analysis of the 2001 Renal Tumour Study Group-International Society of Paediatric Oncology Wilms’ tumour protocol database. Lancet Oncol 2018;19:1072-81. [Crossref] [PubMed]
- Dome JS, Graf N, Geller JI, et al. Advances in wilms tumor treatment and biology: Progress through international collaboration. J Clin Oncol 2015;33:2999-3007. [Crossref] [PubMed]
- Dome JS, Fernandez C V, Mullen EA, et al. Children’s Oncology Group’s 2013 blueprint for research: Renal tumors. Pediatr Blood Cancer 2013;60:994-1000. [Crossref] [PubMed]
- Dome JS, Perlman EJ, Graf N. Risk stratification for wilms tumor: current approach and future directions. Am Soc Clin Oncol Educ Book 2014;215-23. [Crossref] [PubMed]
- Pritchard-Jones K, Kelsey A, Vujanic G, et al. Older age is an adverse prognostic factor in stage I, favorable histology Wilms’ tumor treated with vincristine monochemotherapy: A study by the United Kingdom Children’s Cancer Study Group, Wilm’s Tumor Working Group. J Clin Oncol 2003;21:3269-75. [Crossref] [PubMed]
- Kaste SC, Dome JS, Babyn PS, et al. Wilms tumour: prognostic factors, staging, therapy and late effects. Pediatr Radiol 2008;38:2-17. [Crossref] [PubMed]
- D’Angelo P, Di Cataldo A, Terenziani M, et al. Factors possibly affecting prognosis in children with Wilms’ tumor diagnosed before 24 months of age: A report from the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) Wilms Tumor Working Group. Pediatr Blood Cancer 2017;64:e26644 [Crossref] [PubMed]
- Ehrlich PF, Anderson JR, Ritchey ML, et al. Clinicopathologic findings predictive of relapse in children with stage III favorable-histology wilms tumor. J Clin Oncol 2013;31:1196-201. [Crossref] [PubMed]
- Fernandez CV, Mullen EA, Chi YY, et al. Outcome and prognostic factors in stage III favorable-Histology wilms tumor: A report from the children’s oncology group study AREN0532. J Clin Oncol 2018;36:254-61. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. XLincRNAs: Genomics, evolution, and mechanisms. Cell 2013;154:26. [Crossref] [PubMed]
- Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers 2017;2017:7243968 [Crossref] [PubMed]
- Clark MB, Mercer TR, Bussotti G, et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods 2015;12:339-42. [Crossref] [PubMed]
- Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015;28:529-40. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- Ji Q, Liu X, Fu X, et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/β-catenin signal pathway. PLoS One 2013;8:e78700 [Crossref] [PubMed]
- Qiu MT, Hu JW, Yin R, et al. Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol 2013;34:613-20. [Crossref] [PubMed]
- Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37. [Crossref] [PubMed]
- Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers 2016;2016:9085195 [Crossref] [PubMed]
- de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res 2002;62:2695-8. [PubMed]
- Luo JH, Ren B, Keryanov S, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 2006;44:1012-24. [Crossref] [PubMed]
- Zhang Y, Shields T, Crenshaw T, et al. Imprinting of human H19: Allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am J Hum Genet 1993;53:113-24. [PubMed]
- Bartonicek N, Maag JLV, Dinger ME. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol Cancer 2016;15:43. [Crossref] [PubMed]
- Zhu S, Fu W, Zhang L, et al. LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif 2018;51:e12416 [Crossref]
- Zhao XS, Tao N, Zhang C, et al. Long noncoding RNA MIAT acts as an oncogene in Wilms’ tumor through regulation of DGCR8. Eur Rev Med Pharmacol Sci 2019;23:10257-63. [PubMed]
- Zhang J, Hou T, Qi X, et al. SOX21-AS1 is associated with clinical stage and regulates cell proliferation in nephroblastoma. Biosci Rep 2019;39:BSR20190602 [Crossref] [PubMed]
- Zhu KR, Sun QF, Zhang YQ. Long non-coding RNA LINP1 induces tumorigenesis of Wilms’ tumor by affecting Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2019;23:5691-8. [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773-82. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Frankish A, Diekhans M, Ferreira AM, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766-73. [Crossref] [PubMed]
- Simon N, Friedman J, Hastie T, et al. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw 2011;39:1-13. [Crossref] [PubMed]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1-22. [Crossref] [PubMed]
- Blanche P, Dartigues J-F, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 2013;32:5381-97. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Elkin EB, et al. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak 2008;8:53. [Crossref] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808-15. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Furumichi M, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Res 2019;47:D590-5. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes - Release 72.1, December 1, 2014 . Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci 2019;28:1947-51. [Crossref] [PubMed]
- Tang F, Zhang H, Lu Z, et al. Prognostic Factors and Nomograms to Predict Overall and Cancer-Specific Survival for Children with Wilms’ Tumor. Dis Markers 2019;2019:1092769 [Crossref] [PubMed]
- Grundy PE, Breslow NE, Li S, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: A report from the National Wilms Tumor Study Group. J Clin Oncol 2005;23:7312-21. [Crossref] [PubMed]
- Lu YJ, Hing S, Williams R, et al. Chromosome 1q expression profiling and relapse in Wilms’ tumour. Lancet 2002;360:385-6. [Crossref] [PubMed]
- Hing S, Lu YJ, Summersgill B, et al. Gain of 1q is associated with adverse outcome in favorable histology Wilms’ tumors. Am J Pathol 2001;158:393-8. [Crossref] [PubMed]
- Guo F, Zhang LJ, Liu W, et al. Astrocyte elevated gene-1 overexpression in histologically favorable Wilms tumor is related to poor prognosis. J Pediatr Urol 2014;10:317-23. [Crossref] [PubMed]
- Gong Y, Zou B, Chen J, et al. Potential five-microRNA signature model for the prediction of prognosis in patients with wilms tumor. Med Sci Monit 2019;25:5435-44. [Crossref] [PubMed]
- Gu Y, Feng C, Liu T, et al. The downregulation of lncRNA EMX2OS might independently predict shorter recurrence-free survival of classical papillary thyroid cancer. PLoS One 2018;13:e0209338 [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: More than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
(English Language Editors: J. Jones and J. Chapnick)


