
A novel frameshift mutation in the FGD1 gene causing Aarskog-Scott syndrome patient with hypogonadism: a case report
Introduction
Aarskog syndrome or Aarskog-Scott syndrome (AAS), also called Faciodigitogenital syndrome, was first described in 1970 (1). It is characterized by abnormal facial features (hypertelorism, wide philtrum, round face), short stature, genital anomalies, and skeletal dysplasia. There are two to three new patients with a proven mutation in the FGD1 gene are diagnosed every year, with estimated incidence at around 1/25,000 (2). The growth defects associated with AAS may be present prenatally, and 35% of infants with AAS do not survive their first year due to feeding difficulties and reflux esophagitis. Early signs of mild growth defects appear between ages 1–3, and clinical symptoms became more obvious with age. At present, the disease can only be treated symptomatically.
The clinical manifestations of AAS are very complex, involving almost all systems of the body, with phenotypes and severity varying greatly among individuals (3). Currently, it remains difficult to obtain a definitive clinical diagnosis of ASS due to this complexity. At present, clinical diagnoses do not adequately differentiate AAS from several other congenital diseases such as autosomal dominant inherited forms of Robinow syndrome, Noonan syndrome, pseudohypoparathyroidism, Silver-Russel, and SHORT syndrome (4). It is therefore essential to establish a fast and accurate diagnostic method.
AAS is most commonly transmitted in an X-linked recessive manner through inheritance of the mutations in FGD1 (5). The FGD1 protein is a guanosine exchange factor, which activates the GTPase cell division control protein 42 (Cdc42) by exchanging GDP for GTP; the activated Cdc42 in turn targets downstream intracellular signaling pathways that participate in development processes, such as bone development (6), or maintenance of the extracellular matrix (thereby affecting cell migration and vascular remodeling). Pathogenic mutations that cause loss of FGD1 protein function can result in blocked Cdc42 signaling, and subsequently affect the development of bones and other tissues, ultimately leading to various clinical manifestations of AAS. AAS can also be inherited in an autosomal dominant or an autosomal recessive mode (7,8). Notably, different modes of inheritance have been reported even with one pedigree (9).
This study analyzed a case of AAS diagnosed in our hospital with the hemizygous mutation c.500delA in exon 3 of FGD1 (transcript number: NM_004463.2). A review of the literature confirmed that this pathogenic AAS-causative mutation has not yet been reported.
We present the following case in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/tp-21-26).
Case presentation
The patient, an 11-year and 5-month-old boy, was admitted to the Capital Institute of Pediatrics due to complications associated with a previous orchiopexy for bilateral cryptorchidism. He was born with full-term delivery, birth weight 2,500 g, birth length 48 cm, and without suffocation after birth, and his parents are non-consanguineous. There is no family history of genetic disease, and both parents are in good health and have no special facial features. The father is 168 cm and the mother is 155 cm in height, respectively. Almost 5 years ago, the boy was found to have empty scrotum on both sides and testicles were not touchable. Orchiopexy for bilateral cryptorchidism was performed at the Second Hospital of Qinghai Province. Prior to the second surgery, the child was found the both testicles retreated and the left testicular atrophy. The right testicle can be touched on the right groin, with 2.7×0.9×0.8 cm approximately. The left atrophied testicle was on the left groin, which was difficulted to touch. The texture of the testicles on both sides was hard.
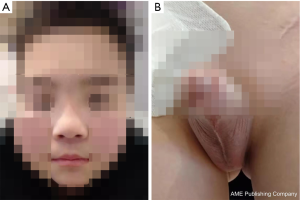
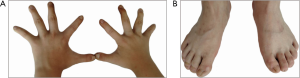
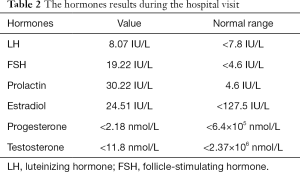
The boy’s body weight was 30 kg (<P3) and a height of 133.8 cm (<P3, –3SD) at the time he was admitted to the hospital at age of 11. The patient exhibited several features characteristic of abnormal genital development, gonadal dysplasia, and clinical syndromes related to gonads. Notably, we observed round face, hypertelorism, broad nasal bridge and curved linear dimple below the lower lip (Figure 1A), a typical scrotal fold and repaired bilateral cryptorchidism (Figure 1B), as well as brachydactyly, joint laxity (Table 1), broad and flat feet, and finger joint hyperextensibility (Figure 2). Although the blood test results for the liver, kidney, blood glucose, or electrolytes were all in normal range, the results of six tests for sex hormones were in consistent with male hypergonadotropin hypogonadism’s characteristics with the decrease of testosterone and the increase of both follicle-stimulating hormone (FSH) and luteinizing hormone (LH) (Table 2).

Full table
Full table
This current study was approved by the Ethics Committee of the Children’s Hospital of Capital Institute of Pediatrics (No. SHERLL2019025) and informed consent was obtained from the patient’s parents. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013).
Methods
WES and data analysis
Genomic DNA was extracted from blood samples provided by the patient and his parents to construct a genomic library. Then whole-exome probes were utilized for capture and enrichment, quality control of the enriched library, and sequencing with a high-throughput sequencer (Illumina). Sequencing data undergoes strict quality control, and sequence comparison was performed after confirming that it was qualified. Then GATK software was used to detect variant sites (10). Based on self-developed software, public databases such as Clinvar, ExAC, 1000Genome, Human Gene Mutation Database (HGMD), and self-built Chinese local database were called for functional annotation. The variants were graded according to the guidelines of American College of Medical Genetics and Genomics (ACMG) during the annotation process.
Sanger sequencing
The FGD1 primers were designed by using UCSC ExonPrimer online software (http://genome.ucsc.edu/index.html) (11). The primers for exon 3 of the FGD1 gene were: Forward, 5'-TTGGCTGTGGCAACATGACT-3' and reverse, 5'-CCATGTTAGCCAGGCTCAAATC-3'. Polymerase chain reaction (PCR) was used to amplify both the exons and exon-intron boundaries. ABI3730XL sequencer was used for DNA sequencing (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and then analyzed with Mutation Surveyor® DNA Variant Analysis Software (version 4.0.4; SoftGenetics, LLC) (12).
In silico analysis of the function of the identified variant
To evaluate the pathogenicity of the variant in the FGD1 gene, we used some tools in silico to analyze it. MultAlin online software was used to analyze the function of the identified variant, which was used to indicate species conservation analysis. Since the PSIPRED (PSI-blast based secondary structure prediction) 4.0.1 server is a rigorous cross-validation technique evaluation method that simply and accurately predicts protein secondary structure (13), it was used in current study to define the related sequences and constrict a position-specific scoring matrix. To generate the sequence profile, the PSI-BLAST was performed and then normalized by PSIPRED. The prediction of an initial secondary structure was made by first neural network, and then a second neural network was used to filter the structure predicted by the first network. And then, PSIPRED was used to predict the secondary structure with the highest score.
Results
Results of the WES
The FGD1 gene (transcript number: NM_004463.2) of this patient has a hemizygous mutation of c.500delA in exon 3 (the 500th nucleotide A in the coding region of the FGD1 gene is deleted), which leads the frameshift variant occurred after tyrosine at position 167 (p.Y167fs).
Results of Sanger sequencing
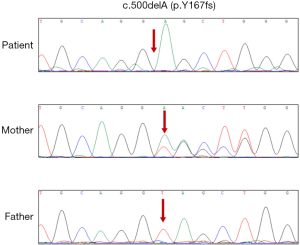
The subsequent Sanger sequencing confirmed the presence of this variant in the patient. Besides, Sanger sequencing results revealed that the patient’s mother, but not the father, to be heterozygous for the same mutation (Figure 3).
Analysis of the function of the identified variant
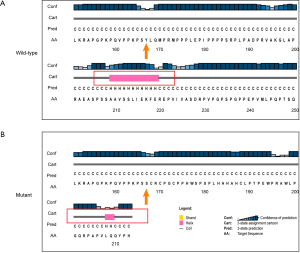
Results from the sequence alignment analysis indicated that the amino acid was highly evolutionarily conserved after codon 167 across species ranging from Homo sapiens to Mus musculus (Figure 4). This frameshift deletion is predicted to shift the reading frame in a way that translates to 47 erroneous amino acids followed by a premature stop codon at position 213 (Figure 5). This suggests that the p.Y167fs completely destroyed the primary structure of FGD1, which was very rarely found in the general population. We compared the secondary structures of normal and mutant proteins using computer-based calculations. This frameshift mutation resulted in the conversion of the α-helix (pink cylinder) in the wild type protein to a random coil and changed the length of the α-helix (Figure 5). The p.Y167 frameshift mutation changed the structural stability of the α-helix and therefore disrupted the normal protein structure.

Discussion
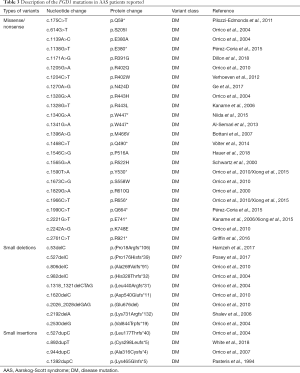
Although mutations in the FGD1 gene are so far the only identified genetic cause of AAS, only about 20% of Aarskog families have been found to carry pathogenic mutations in FGD1 located in Xp11.21 (14). Thus, there are currently less than 100 children with definite genetic diagnoses of AAS worldwide. A search of PubMed showed that only 37 relevant mutations, all associated with AAS, have been confirmed using molecular testing according to the HGMD (see Table 3).
Full table
The c.500delA mutation of FGD1 identified in this patient is hemizygous, thus conforming to the X-linked recessive inheritance mode of AAS. Moreover, although no study has yet reported this mutation as a cause of AAS, other, similar frameshift mutations in FGD1 are recorded in the clinVar database, including the pathogenic or likely pathogenic mutations at amino acid residues 176, 177, 298, 316, 705, and 713 (all of which occur after Tyr167). This mutation appears to be extremely rare, and no records of its occurrence are found in the ExAC or Thousand Genomes Databases.
By performing sequence alignment analysis, our results demonstrated that the amino acids were highly evolutionarily conserved in the highly conserved region after the codon 167, indicating a possibility of important function. The frameshift deletion from this patient shifted the reading frame which translates to 47 erroneous amino acids followed by a premature stop codon, which completely destroyed the primary structure of FGD1. In addition, the secondary structure of the FGD1 protein suggested that the frameshift mutations resulted in the conversion of the α-helix to a random coil and changed the length of the α-helix, leading to a change in the structural stability of the α-helix and therefore impaired the normal protein structure. The damage in the secondary structure changed in the RhoGEF gene domain and ultimately influenced the function of the FGD1 protein, which means that it is a disease-causing variant. Based on the above evidence, and according to the ACMG guidelines, the grade of the mutation is confirmed as a pathogenic variant. Pedigree validation indicated that the c.500delA mutation was maternally inherited, while the father’s single copy of FGD1 showed no c.500delA mutation.
Due to this pattern of X-linked inheritance, AAS patients are most frequently male, and the clinical manifestations of facial abnormalities, skeletal deformities, and abnormal external genitalia comprise a characteristic triad of diagnostic features (2). AAS can also affect neurodevelopment, and some children may be accompanied by mild intellectual disability and attention deficit hyperactivity disorder, requiring intervention under the guidance of a neurologist (15,16). Failure-to-thrive is a common cause of treatment for this disease. The growth hormone stimulation test is often normal in children with AAS, but patients could show delayed puberty (17). Here, we are reporting a clinic case with the molecular analysis from a patient with AAS and his family reveals that a novel FGD1 mutation causes the repaired bilateral cryptorchidism associated with hypogonadism. The hormone examination results are consistent with male hypergonadotropin hypogonadism’s characteristics with the decrease of testosterone and the increase of both FSH and LH. The anomalies of the testis and hormone levels are severe, and may lead to probability of infertility (18). The clinical features accompanied this case, notably the short stature (<P3, –3SD) and faciogenital dysplasia, such as round face, hypertelorism, typical scrotal fold, and joint laxity, strongly suggested the AAS diagnostic triad.
Orrico et al. identified 24 cases of pathogenic mutations in a family study covering 110 suspected cases of the disease, with a detection rate of 21.8% (19). This low detection rate may also be at least partially attributable to the high similarity in clinical manifestations between AAS and several other syndromes. As prenatal diagnostic techniques improve, so should the timeliness of intervention for many genetic diseases with poor prognosis. However, AAS differs from other serious or fatal congenital diseases in that the clinical manifestations caused by mutations in FGD1 can be highly heterogeneous, even within a single family. Given this heterogeneity and subsequent unpredictability of later clinical symptoms, this disease can only be treated through a multidisciplinary cooperative symptom-based approach. Hypospadias, inguinal or umbilical hernias, cryptorchidism, and severe craniofacial deformities can be resolved by surgery. By contrast, the therapeutic effects of growth hormone have only been reported in preliminary studies, though the apparently positive effects on body development and final height are promising (20). However, due to the low number of cases and the lack of randomized controlled studies, more data on the effects of these treatments in children are needed to draw reliable conclusions.
Thus, our study shows that frameshifts disrupting the RhoGEF gene domain of FGD1 represent the most prevalent causal mutations underlying AAS and expand the phenotypic and mutational spectra of this disease. Improved understanding of the phenotypic and pathological heterogeneity accompanying FGD1 mutation can significantly enhance the clinical prognostic capabilities in the future and aid genetic counseling for AAS patients.
Acknowledgments
We thank the proband and his family for participating in this study.
Funding: This work was partly supported by the Key Medical Specialty Development Plan of Beijing Hospital Administration (grant number ZYLX201825), and the Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority (grant number XTYB201823).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/tp-21-26
Peer Review File: Available at http://dx.doi.org/10.21037/tp-21-26
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-21-26). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the Children’s Hospital of Capital Institute of Pediatrics (No. SHERLL2019025) and informed consent was taken from the patient’s parents. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aarskog D. A familial syndrome of short stature associated with facial dysplasia and genital anomalies. J Pediatr 1970;77:856-61. [Crossref] [PubMed]
- Orrico A, Galli L, Clayton-Smith J, et al. Clinical utility gene card for: Aarskog-Scott syndrome (faciogenital dysplasia). Eur J Hum Genet 2011; [Crossref] [PubMed]
- Orrico A, Galli L, Cavaliere ML, et al. Phenotypic and molecular characterisation of the Aarskog-Scott syndrome: a survey of the clinical variability in light of FGD1 mutation analysis in 46 patients. Eur J Hum Genet 2004;12:16-23. [Crossref] [PubMed]
- Parıltay E, Hazan F, Ataman E, et al. A novel splice site mutation of FGD1 gene in an Aarskog-Scott syndrome patient with a large anterior fontanel. J Pediatr Endocrinol Metab 2016;29:1111-4. [Crossref] [PubMed]
- Völter C, Martínez R, Hagen R, et al. Aarskog-Scott syndrome: a novel mutation in the FGD1 gene associated with severe craniofacial dysplasia. Eur J Pediatr 2014;173:1373-6. [Crossref] [PubMed]
- Pasteris NG, Cadle A, Logie LJ, et al. Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 1994;79:669-78. [Crossref] [PubMed]
- van de Vooren MJ, Niermeijer MF, Hoogeboom AJ. The Aarskog syndrome in a large family, suggestive for autosomal dominant inheritance. Clin Genet 1983;24:439-45. [Crossref] [PubMed]
- Teebi AS, Naguib KK, Al-Awadi S, et al. New autosomal recessive faciodigitogenital syndrome. J Med Genet 1988;25:400-6. [Crossref] [PubMed]
- Xu M, Qi M, Zhou H, et al. Familial syndrome resembling Aarskog syndrome. Am J Med Genet A 2010;152A:2017-22. [Crossref] [PubMed]
- Walker MA, Pedamallu CS, Ojesina AI, et al. GATK PathSeq: a customizable computational tool for the discovery and identification of microbial sequences in libraries from eukaryotic hosts. Bioinformatics 2018;34:4287-9. [Crossref] [PubMed]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996-1006. [Crossref] [PubMed]
- Kénémé B, Sembène M. GTn repeat microsatellite instability in uterine fibroids. Front Genet 2019;10:810. [Crossref] [PubMed]
- Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res 2019;47:W402-7 [Crossref] [PubMed]
- Verhoeven WM, Egger JI, Hoogeboom AJ. X-linked Aarskog syndrome: report on a novel FGD1 gene mutation. Executive dysfunction as part of the behavioural phenotype. Genet Couns 2012;23:157-67. [PubMed]
- Kaname T, Yanagi K, Okamoto N, et al. Neurobehavioral disorders in patients with Aarskog-Scott syndrome affected by novel FGD1 mutations. Am J Med Genet A 2006;140:1331-2. [Crossref] [PubMed]
- Orrico A, Galli L, Buoni S, et al. Attention-deficit/hyperactivity disorder (ADHD) and variable clinical expression of Aarskog-Scott syndrome due to a novel FGD1 gene mutation (R408Q). Am J Med Genet A 2005;135:99-102. [Crossref] [PubMed]
- Darendeliler F, Larsson P, Neyzi O, et al. Growth hormone treatment in Aarskog syndrome: analysis of the KIGS (Pharmacia International Growth Database) data. J Pediatr Endocrinol Metab 2003;16:1137-42. [Crossref] [PubMed]
- Bonora G, Baronciani D, Perletti L, et al. Aarskog syndrome. Description of a case with significant anomalies of the gonads. Pediatr Med Chir 1985;7:909-11. [PubMed]
- Orrico A, Galli L, Faivre L, et al. Aarskog-Scott syndrome: clinical update and report of nine novel mutations of the FGD1 gene. Am J Med Genet A 2010;152A:313-8. [Crossref] [PubMed]
- Şıklar Z, Berberoğlu M. Syndromic disorders with short stature. J Clin Res Pediatr Endocrinol 2014;6:1-8. [Crossref] [PubMed]


