
Comprehensive analysis of clinical spectrum and genotype associations in Chinese and literature reported KBG syndrome
Introduction
The KBG syndrome (MIM #148050), named after the first reported families (1), is an autosomal dominant disease currently known to be caused by either an ANKRD11 gene mutation (2) or a microdeletion in chromosome 16q24.3 including the ANKRD11 gene (3-5). The KBG syndrome is characterized by macrodontia, intellectual disability (ID), short stature (SS), delayed bone age, hand findings, costovertebral anomalies, characteristic facial dysmorphism, and the presence of a first-degree relative with the syndrome (6).
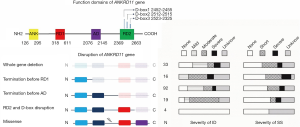
The ANKRD11 gene encodes an ankyrin repeat-containing protein (ANKRD11 or previous ANCO-1), which is indispensable in neuron proliferation (7). Earlier in vitro study showed that ANKRD11 protein acts as a transcriptional repressor (8) and functions through its two transcriptional repression domains (RDs: RD1, aa 318–611; and RD2, aa 2,369–2,663) and one activation domain (AD, aa 1,851–2,145) (9). In addition, the RD2 domain regulates the activity of the ANKRD11 protein and maintains the protein’s appropriate degradation (10).
In vivo studies showed that either ANKRD11 gene knockdown or the accumulation of truncated ANKRD11 protein can result in the KBG phenotype (10,11). Recent advances in genetic analysis have allowed the identification of patients with extremely mild symptoms of the KBG syndrome (12-16). Patients with the KBG syndrome, who commonly seek medical consultation and treatment owing to malformations, ID, or SS, demonstrate great variations in these traits, and proper clinical diagnosis is frequently difficult (17-19).
Studies focusing on the relationship between genotype and phenotype in the KBG syndrome remain scarce. Here we report three new genetically diagnosed Chinese KBG patients and further analyze the underlying genotype-phenotype relationship in combination with 186 cases reported in the literature.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tp-20-385).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Children’s Hospital of Fudan University (No. 2017237), and written informed consent was obtained from the patient.
Patients and diagnose criteria
Three unrelated probands were clinical diagnosed as having KBG symptoms using the diagnostic criteria proposed by Skjei et al. (6). All three patients presented four of the following major clinical features: macrodontia, ID, SS, delayed bone age, hand findings, costovertebral anomalies, characteristic facial dysmorphism, and the presence of a first-degree relative with the syndrome. The family members were invited for further clinical investigations and genetic tests after acquiring informed consent from all parents.
Whole exome sequencing (WES) analysis
WES was performed in each family by WuXi NextCODE Genomics, Shanghai, China (CLIA Lab ID: 99D2064856) (20). Briefly, exome capture was performed using the Agilent SureSelect Human All Exon V5, Illumina TruSeq Rapid PE Cluster, and SBS kits (Agilent Technologies, Santa Clara, CA, USA). Illumina HiSeq provided the 2000/2500 platform for WES. Reads alignment were referenced to the human genome sequence (http://genome.ucsc.edu) through the Burrows-Wheeler Aligner v.0.6.2. Duplicate paired-end reads were marked with Picard v.1.55 (https://broadinstitute.github.io/picard/). The base quality score recalibration, indel realignment, and variant discovery were performed by the Genome Analysis Toolkit v.2.3-9. Variants were annotated using a pipeline developed in-house (20) and filtered in the gnomAD, Exome Variant Server, dbSNP databases, and Exome Aggregation Consortium.
Systematic literature review
Articles in the databases Ovid MEDLINE and Web of Knowledge (2011 June to 2019 Aug) were searched using the following terms: ankyrin repeat domain 11, ANKRD11, ankyrin repeats containing cofactor, ANCO-1, 16q24.3, and KBG. OMIM and ClinVar databases were also searched for mutations in the ANKRD11 gene and related publications in Human Gene Mutation Database (HGMD). In addition, a manual search for references of initially identified articles was carried out. The clinical data [gender, family of origin, age at diagnosis, and types of ANKRD11 gene mutation (in reference to ENST00000378330.7)] or 16q23.4 microdeletion (aligned to GRCh38 coordinates using the UCSC liftover tool) of reported cases in each publication were collected. Literature selection and data extraction were performed independently by two investigators (Q Li and C Sun). The data were then reviewed by a third investigator (F Luo) and the conflicts were resolved by consensus.
Intelligence quotient (IQ) and body height classification
The degree of ID was determined based on the description of intelligence estimation in each report (by the measurement of either IQ or equivalent such as developmental quotient derived from milestones, school attendance, levels of special educational need, or basis of daily skills) and categorized according to Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) stages of ID (i.e., mild: IQ 50–69; moderate: IQ 36–49; severe: IQ 20–35) (21), The degree of SS was categorized according to cutoffs commonly used in practice [i.e., SS: height less than 3rd percentile or −2 standard deviation scores (SDS); severe SS: height less than 1st percentile or −3 SDS] per description in each report (22).
ANKRD11 gene variant grouping
Based on the functional domains of the ANKRD11 gene, variants were further grouped based on different types and positions: (I) deletion of the whole ANKRD11 gene; (II) premature stop before the RD1 domain (deletion of RD1, AD, and RD2); (III) premature stop located between the RD1 and AD domains (deletion of AD and RD2); (IV) premature stop truncating the RD2 domain or its D-box (deletion of RD2 alone); and (V) missense mutations.
Statistic analysis
Data were analyzed using R software (version 3.6.1, www.r-project.org). The quantitative data were compared between groups using Fisher’s exact test. The frequencies trend was analyzed by Cochran-Armitage test. A P value <0.05 was considered to be statistically significant.
Results
Patient 1
The patient was an 8-year-old girl born to non-consanguineous healthy parents, and was referred to our clinic owing to postnatal growth retardation. She was born slightly small with a birth weight of 2.4 kg and length of 48 cm. She started to walk around 1.5 years of age and started to speak around 2 years of age. Upon her presentation at age of 8 years, she had a height of 110 cm (−3.4 SD) and weight of 17.7 kg (−2.5 SD). She had triangular face, low posterior hairline, arched eyebrows, long eye lashes, broad nasal low bridge, flat philtrum, thin upper lip, wide upper central incisors and prominent anteverted ears, short fingers with fifth finger clinodactyly (Figure S1A,B). A skeletal survey revealed 1-year delayed bone age and intoeing (pigeon toes). Her IQ was 56. The patient was found to carry de novo mutation (c.6792dupC, p. P2271Pfs*8) in ANKRD11 gene (Figure S1C). Examination of both parents revealed normal phenotypes. Her father (height 174 cm, +0.32 SD) was an engineer, and her mother (height 162 cm, +0.38 SD) was unemployed and stayed at home.
Patient 2
The patient 2, a 2-year-old boy, born to non-consanguineous healthy parents, was referred to our clinic owing to postnatal growth retardation. His birth weight was 2.6 kg and the length was 48 cm. He started to walk around 1.5 years of age. His height was 85.5 cm (−3.1 SD) and body weight was 12 kg (−0.4 SD) when accepted at the hospital. Clinical examination revealed facial dysmorphisms, including round face, tall forehead, long eyelashes and prominent anteverted ears, marked enamel hypoplasia (Figure S1D) and fifth finger clinodactyly (Figure S1E). A novel nonsense variant (c.7407dupC, p. P2530Rfs*61) (Figure S1C) was identified, which was inherited from his mother. His mother (height 148 cm; −2.2 SD) was not able to complete high school and currently self-employed in small ecommerce business; She had round face, wide upper central incisors, and prominent anteverted ears. His father (height 169 cm; −0.65 SD) had normal phenotypes, and his genetic tests were normal.
Patient 3
The third patient was an 8.1-year-old boy from a non-consanguineous family, and was presented to us for the evaluation of his SS. At his presentation, he was 116.9 cm (−3.5 SD) tall and weighed 23.4 kg (−1 SD). He had a symmetrical face, pointed chin and bilateral ptosis. His lips were full and thick. The upper central permanent incisors were large. His bone age was 2 years delayed. The boy was noticed to have severe bilateral strabismus since 4 years of age. The boy was reported to be healthy despite progressive postnatal growth failure with a gradual decline in height. He is currently studying in normal primary school (3rd year) and his intellectual performance was normal in his class. He was born full term after uncomplicated pregnancy with normal measurements (length 50 cm, weight 3.0 kg). The genetic analysis of the family showed that he carried a novel (c. G3046A, p. D1016N) mutation in the ANKRD11 gene (Figure S1C). His father had SS (height: 160 cm, −2.1 SDS) and his mother (height 163 cm, +0.67 SD) had normal phenotypes.
Genetic characteristics of KBG syndrome
The systematic searches yielded 186 patients from 155 families presented as KBG syndrome with ANKRD11 gene pathological variants or 16q24.3 microdeletions in the literature (Supplementary data, available at https://cdn.amegroups.cn/static/public/tp-20-385-1.xlsx). Among those patients, females were 46% (85/186). The median age of the patients receiving genetic diagnosis was 11.6 years (IQR, 6.5–19.0 years; range, 0.9–66.0 years, based on 168 available data). A reported family history of KBG syndrome was identified in 30 (24.8%) patients, 30% patients presented moderate to severe ID, and 53% patients had SS (Table 1).
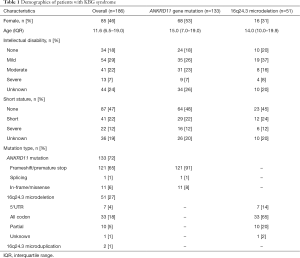
Full table
Among the 186 patients, 71.5% (133/186) patients carried heterozygous ANKRD11 gene pathological variants, 27.4% (51/186) patients carried heterozygous 16q24.3 microdeletions with disrupted ANKRD11 gene, 1.1% (2/186) carried 16q24.3 microduplications (Table 1). In the patients who carried ANKRD11 gene mutation, the majority of patients (91%) carried nonsense or frameshift variants leading to a premature termination of the protein (Table 1). There were frequent pathological variants such as p.Lys635Glnfs*26 (21 patients in 16 families), p.Glu461Glnfs*48 (4 patients in 3 families), p.Glu1075Glyfs*242 (2 patients in 2 families), and deletions between c.2395 and c.2412 (9 patients in 7 families, Table S1). Most of the 16q24.3 microdeletions in those patients (65%) disrupted the whole ANKRD11 gene, resulting in complete allelic absence (Table 1).
Genotype and phenotype association in the KBG syndrome
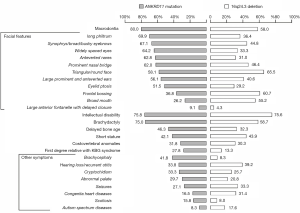
Patients with ANKRD11 gene variants had higher prevalence of macrodontia (80% vs. 58% P=0.002) and some facial features, including long philtrum (69.9% vs. 36.4% P=0.001), abnormal eyebrows (67.1% vs. 44.8%, P=0.04), widely spaced eyes (64.2% vs. 33.3%, P=0.04), anteverted nares (62.8% vs. 31.0%, P=0.003), and eyelid ptosis (51.5% vs. 29.2%, P=0.04) than those with a 16q24.3 microdeletion, as well as such skeletal presentations as brachydactyly (75.0% vs. 58.7%, P=0.04) and brachycephaly (41.8% vs. 8.3%, P=0.001). However, 16q24.3 microdeletion was associated with an increased risk of heart diseases compared with ANKRD11 gene mutation (31.4% vs. 16.5%, P=0.02) and autism spectrum disorder (ASD) (18.7% vs. 8.3%, P=0.04), as well as frontal bossing (36.8% vs. 64.2%, P=0.04). Other features were similar among the two groups (Figure 1). No difference was detected in disease severity in ID and SS between ANKRD11 gene pathological variants and 16q24.3 microdeletions (Table 1).
As shown in Table 2, the frequency of individuals with moderate to severe ID was significantly lower among KBG patients carrying truncating variants between RD1 and AD than those carrying pathological variants with a premature stop before the RD1 domain (24% vs. 50%, P=0.04) and a premature stop truncating the RD2 domain or its D-box (24% vs. 58%, P=0.015). Although terminations close to the C-terminal of the ANKRD11 protein tended to have less severe SS (Figure 2), neither the difference nor trend in the severity of SS reached statistically significant among different ANKRD11 gene mutation types (Table S2).
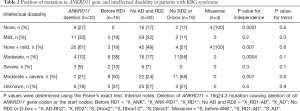
Full table
Discussion
Patients with KBG syndrome may present a wide spectrum of clinical features, such as malformations, ID, or SS (12-14). Together with ours and the literature reported KBG syndrome cases, we found that the severity of ID in KBG syndrome was associated with the position of the ANKRD11 gene mutation. The ID in patients with KBG syndrome was significantly more severe among KBG patients carrying pathological variants disrupting either all functional domains or the RD2 domain alone than those carrying truncated variants located between the RD1 and AD domains. To our knowledge, this is the first study that shows the importance of the intact repression function of the ANKRD11 protein for the development of normal intelligence by clinical data.
Our analysis revealed that KBG syndrome was predominantly caused by heterozygous ANKRD11 gene pathological variants (71.5%, 133/186). Microdeletions or microduplications were responsible for the remaining causes. Previous studies showed that there is no difference in the degree of ID between ANKRD11 gene deletion and mutation patients (3,18). However, our current data analysis demonstrated that patients with pathological variants retaining RD1 alone had milder ID than those retaining RD1 and AD simultaneously. This could be explained by Ka et al. previous findings, in which the repression effect of RD1 is weaker than the ability to activate transcription of AD, which promotes acetylation of epigenetic molecules such as p53 and histone H3 to repress neuron differentiation (11). In addition, an in vivo study showed that disrupting the RD2 domain results in accumulation of abnormal proteins that may further aggravate the destruction of neurons (10). A recent study has demonstrated that decreased ANKRD11 gene expression is associated with abnormal neuron differentiation (3), and our results showed that pathological variants preserving only the RD1 had similar intelligence reduction when compared to the full ANKRD11 gene deletion, suggesting that haploinsufficiency of ANKRD11 protein may be the mechanism in the autosomal dominant KBG syndrome. In the published literature, we found that four patients with missense mutations of ANKRD11 gene presented normal intelligence (10,13,23), and the same result was found for our patient 3.
Beyond ID, our current study also revealed other genotype-phenotype associations for different ANKRD11 gene defects. In the ANKRD11 gene mutation group, the detailed manifestation of facial changes, such as macrodontia, long philtrum, abnormal eyebrows, widely spaced eyes, anteverted nares, eyelid ptosis, brachydactyly, and brachycephaly were frequently observed, whereas 16q24.3 microdeletion was more frequently associated with prominent forehead, congenital heart diseases, and autism spectrum diseases. This observation is consistent with the findings of Willemsen et al. (3) and Isrie et al. (24) studies. Willemsen et al. (3) suggested that deletion of the 16q24.3 region should be recognized as a novel and distinctive microdeletion syndrome, because its features differ from those of the syndrome caused by ANKRD11 gene pathological variants. Although our three patients manifested ID or language delay, they all lacked signs of ASD. These three cases provide additional support to the finding that ASD are likely strong features of the KBG syndrome caused by 16q24.3 microdeletion.
Few studies have been conducted to understand the SS status in patients with KBG syndrome, and its detailed molecular mechanism is still unknown. Recent in vivo studies showed that ethyl nitrosourea (ENU)-induced mutation in the ANKRD11 gene presented r reduced body size and bone mass (25), suggesting that this gene plays an important role in regulating linear growth. Although the current data suggested that less disrupted ANKRD11 protein tend to have less severe SS, the statistical insignificance revealed that association between ANKRD11 gene mutation and SS needs to be further investigated by future studies with more subjects. Reynaert et al. (26) reported that the mean SDS of height is −2.56 in children and −2.17 in adults. Similarly, our three patients had severe SS (<−3 SD), and adults’ heights (parents of patients 2 and 3) ranged from −2.1 SD to −2.2 SD. The above results suggest that even without the intervention of growth hormone, children’s adult heights will increase spontaneously to near the normal low limit, thus the growth pattern of KBG syndrome still needs to be further studied in the future.
Our findings showed that patients with ANKRD11 gene variants disrupting RD1 and RD2 or RD2 alone are more likely to have severe ID, in view of their higher risks, an early recognition of KBG syndrome severity subtype can be of great benefits for individual intervention. Although we detected significant differences between the effects of different domain in ANKRD11 protein on intelligence, we cannot exclude the possibility that these effects might have arisen from differences in populations or methodological differences for evaluating ID.
In summary, our study identified three Chinese KBG syndrome patients and two new pathological variants in the ANKRD11 gene. Combined with the literature cases, we found that individuals with ANKRD11 gene variants disrupting their repression function domains are more likely to have more severe ID, which warrants early intervention for the mental development of the patient.
Acknowledgments
The authors are grateful to all family members for their valuable contribution to the present study. The authors also thank all the physicians and nurses for help with management of these patients.
Funding: This study was supported by the National Natural Science Foundation of China (grant No. 81873667).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-385
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-385
Peer Review File: Available at http://dx.doi.org/10.21037/tp-20-385
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-385). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Children’s Hospital (No. 20170237) and written informed consent was obtained from the patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herrmann J, Pallister PD, Tiddy W, et al. The KBG syndrome-a syndrome of short stature, characteristic facies, mental retardation, macrodontia and skeletal anomalies. Birth Defects Orig Artic Ser 1975;11:7-18. [PubMed]
- Sirmaci A, Spiliopoulos M, Brancati F, et al. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am J Hum Genet 2011;89:289-94. [Crossref] [PubMed]
- Willemsen MH, Fernandez BA, Bacino CA, et al. Identification of ANKRD11 and ZNF778 as candidate genes for autism and variable cognitive impairment in the novel 16q24.3 microdeletion syndrome. Eur J Hum Genet 2010;18:429-35. [Crossref] [PubMed]
- Youngs EL, Hellings JA, Butler MG. ANKRD11 gene deletion in a 17-year-old male. Clin Dysmorphol 2011;20:170-1. [Crossref] [PubMed]
- Sacharow S, Li D, Fan YS, et al. Familial 16q24.3 microdeletion involving ANKRD11 causes a KBG-like syndrome. Am J Med Genet A 2012;158A:547-52. [Crossref] [PubMed]
- Skjei KL, Martin MM, Slavotinek AM. KBG syndrome: report of twins, neurological characteristics, and delineation of diagnostic criteria. Am J Med Genet A 2007;143A:292-300. [Crossref] [PubMed]
- Gallagher D, Voronova A, Zander MA, et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev Cell 2015;32:31-42. [Crossref] [PubMed]
- Zhang A, Yeung PL, Li CW, et al. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J Biol Chem 2004;279:33799-805. [Crossref] [PubMed]
- Zhang A, Li CW, Chen JD. Characterization of transcriptional regulatory domains of ankyrin repeat cofactor-1. Biochem Biophys Res Commun 2007;358:1034-40. [Crossref] [PubMed]
- Walz K, Cohen D, Neilsen PM, et al. Characterization of ANKRD11 mutations in humans and mice related to KBG syndrome. Hum Genet 2015;134:181-90. [Crossref] [PubMed]
- Ka M, Kim WY. ANKRD11 associated with intellectual disability and autism regulates dendrite differentiation via the BDNF/TrkB signaling pathway. Neurobiol Dis 2018;111:138-52. [Crossref] [PubMed]
- Ockeloen CW, Willemsen MH, de Munnik S, et al. Further delineation of the KBG syndrome phenotype caused by ANKRD11 aberrations. Eur J Hum Genet 2015;23:1176-85. [Crossref] [PubMed]
- Miyatake S, Okamoto N, Stark Z, et al. ANKRD11 variants cause variable clinical features associated with KBG syndrome and Coffin-Siris-like syndrome. J Hum Genet 2017;62:741-6. [Crossref] [PubMed]
- Scarano E, Tassone M, Graziano C, et al. Novel mutations and unreported clinical features in KBG syndrome. Mol Syndromol 2019;10:130-8. [Crossref] [PubMed]
- Alves RM, Uva P, Veiga MF, et al. Novel ANKRD11 gene mutation in an individual with a mild phenotype of KBG syndrome associated to a GEFS+ phenotypic spectrum: a case report. BMC Med Genet 2019;20:16. [Crossref] [PubMed]
- Crippa M, Rusconi D, Castronovo C, et al. Familial intragenic duplication of ANKRD11 underlying three patients of KBG syndrome. Mol Cytogenet 2015;8:20. [Crossref] [PubMed]
- Kang Y, He D, Li Y, et al. A heterozygous point mutation of the ANKRD11 (c.2579C>T) in a Chinese patient with idiopathic short stature. Mol Genet Genomic Med 2019;7:e988 [Crossref] [PubMed]
- Goldenberg A, Riccardi F, Tessier A. el al. Clinical and molecular findings in 39 patients with KBG syndrome caused by deletion or mutation of ANKRD11. Am J Med Genet A 2016;170:2847-59. [Crossref] [PubMed]
- Lo-Castro A, Brancati F, Digilio MC, et al. Neurobehavioral phenotype observed in KBG syndrome caused by ANKRD11 mutations. Am J Med Genet B Neuropsychiatr Genet 2013;162B:17-23. [Crossref] [PubMed]
- Yang L, Kong Y, Dong X, et al. Clinical and genetic spectrum of a large cohort of children with epilepsy in China. Genet Med 2019;21:564-71. [Crossref] [PubMed]
- Gmitrowicz A, Kucharska A. Developmental disorders in the fourth edition of the American classification: diagnostic and statistical manual of mental disorders (DSM IV -- optional book). Psychiatr Pol 1994;28:509-21.
- Grimberg A, DiVall SA, Polychronakos C, et al. Guidelines for growth hormone and insulin-like growth factor-I treatment in children and adolescents: growth hormone deficiency, idiopathic short stature, and primary insulin-like Growth Factor-I Deficiency. Horm Res Paediatr 2016;86:361-97. [Crossref] [PubMed]
- De Bernardi ML, Ivanovski I, Caraffi SG, et al. Prominent and elongated coccyx, a new manifestation of KBG syndrome associated with novel mutation in ANKRD11. Am J Med Genet A 2018;176:1991-5. [Crossref] [PubMed]
- Isrie M, Hendriks Y, Gielissen N, et al. Haploinsufficiency of ANKRD11 causes mild cognitive impairment, short stature and minor dysmorphisms. Eur J Hum Genet 2012;20:131-3. [Crossref] [PubMed]
- Barbaric I, Perry MJ, Dear TN, et al. An ENU-induced mutation in the Ankrd11 gene results in an osteopenia-like phenotype in the mouse mutant Yoda. Physiol Genomics 2008;32:311-21. [Crossref] [PubMed]
- Reynaert N, Ockeloen CW, Sävendahl L, et al. Short stature in KBG syndrome: first responses to growth hormone treatment. Horm Res Paediatr 2015;83:361-4. [Crossref] [PubMed]


