
Alterations in biochemical markers in adenovirus infection
Introduction
Respiratory infections are very common, particularly in young children (under 5 s). Viruses are responsible for the majority of these infections, and as such viral infection is one of the principal causes of infant mortality (1,2). Most current diagnostic techniques have led to greater diagnostic sensitivity and allow the type of viral infection, and its characteristics, to be identified. Of the more serious respiratory viruses, Adenovirus (HAdV) is one of the most frequently encountered (3). Children with HAdV generally present with prolonged fever and a strong inflammatory response, which is similar to bacterial infections. The gravity of lower respiratory tract infections caused by HAdV has been linked to the infecting genotype, age and the state of the patient’s immune system (4,5).
One of the most frequent actions in daily clinical practice in the context of infections is the analysis of biochemical and/or haematological markers. Biomarkers such as Procalcitonin (PCT) and C-reactive protein (CRP) are used in connection with bacterial infection (6) and PCT has been successfully employed in the management of antibiotic therapy (7). However, recent research data indicates that these tests are not of equal relevance for all types of patients (8) and suggests an association with viral infections in addition to bacterial (9). Establishing the link between specific biochemical markers and respiratory virus infection could have an immediate effect on the management of young children with respiratory infections and help to avoid unnecessary antibiotic treatment (10).
The present study aimed to identify the frequency of respiratory infection caused by HAdV in children presenting with fever and respiratory difficulties, and its relationship with biochemical (PCT and CRP) and haematological biomarkers of infection.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tp-20-333).
Methods
Patients and samples
Over a single epidemiological monitoring season (September 2015–June 2016), the Virology Department at the Hospital Universitario Central de Asturias (HUCA) received 3,441 pharyngeal and nasal pharyngeal exudates from the Paediatric Services department. From among these, 487 samples (293 boys and 194 girls) were selected for further investigation, all belonging to children that had presented in the Emergency Department of the HUCA with fever and/or respiratory compromise and for whom peripheral blood samples were taken to detect biochemical markers (PCT and CRP) and haematological markers (leukocyte formula and count). Eighty of these children were under one year old, 298 were between 1 and 5 years, and 109 between 6 and 14. In terms of their clinical symptoms, 361 (74.1%) had a fever at the time of presenting in the Emergency Department (ED) (this being the principal symptom for 282 children who had had elevated temperature for a number of days, and it was accompanied by respiratory symptoms in 79 of these cases), 146 (30%) had upper respiratory tract infection (URTI) symptoms (pharyngitis, tonsillitis, otitis), 23 (4.7%) had symptoms of lower respiratory tract infection (LRTI, bronchitis, pneumonia), 36 (7.4%) had suspected flu (SF), and in addition, 4 children presented with diarrhoea and 5 with exanthema in addition to respiratory symptoms/fever. In 141 of the 487 children selected for this work, samples were also taken in the ED and sent for analysis to determine whether bacterial infection was present. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and has been approved by Hospital Universitario Central de Asturias Ethical Committee (nº 2020.383). Our study is based on the use of remnants of samples used for routine diagnosis. The retrospective collection of the informed consent belonging to the collection of samples stored by the Mycrobiology Service of HUCA was complicated, so the Ethical Committee was asked to exempt informed.
Virological methods
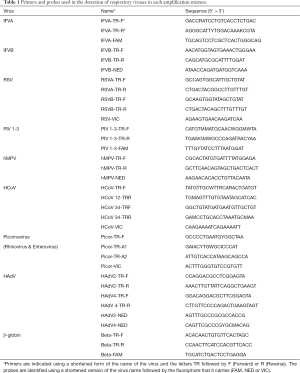
For the virological studies, the pharyngeal and nasopharyngeal exudates were processed following the protocols of the Virology Department of the HUCA. This involved each sample being divided into three aliquots. The first (1 mL) was used to inoculate the cell lines MDCK, LLC-MK2 and MRC-5 (following established conventional protocols). With the second (500 µL), after 24 h, the standard techniques to detect Ag using immunofluorescence were employed to detect monoclonal antibodies to respiratory viruses: Adenovirus (HAdV), Influenza A and B (IFVA, IFVB), respiratory syncytial virus (RSV), Parainfluenza virus (PIV1-3), Enterovirus (ETV) and Metapneumovirus (hMPV) (Sanofi Pasteur, Lyon, France; Dako, Glostrup, Denmark).
From the final aliquot (500 µL), nucleic acids were extracted using automatic extractors (Magnapure or COBAS Ampripep; Roche Diagnostics SL, Mannheim, Germany) following the manufacturer’s instructions. Then, using the purified nucleic acids genomic detection was carried out using 3 different multiple Real-Time qPCR with prior retrotranscription (RT-PCR): one for IFVA, IFVB and RSV; one for PIV, hMPV and Coronavirus (HCoV); and the other for Picornaviruses [ETV, Rhinovirus (HRV)] and which included the detection of human β-globin.
The next step involved the amplification of 5 µL of the extracted genome in a final volume of 10 µL which included TaqMan Fast 1-Step Master Mix, primers (Thermo Fisher Scientific, Waltham, Massachusetts) and MGB Taqman probes marked with the flourophores FAM, VIC and NED, each target depending on the specific virus involved (Table 1). The amplifications were made using the thermal cyclers 7300 and 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, Massachusetts) using a thermal profile of a initial step at 50 °C for 20 s, followed by 40 cycles of 90 °C for 10 s and 60 °C for 35 s.
Full table
Genomic quantification was calculated using the correlation between the Ct of the specific virus involved and the Ct of human β-globin (11). The genomic quantification, or viral load (VL), is thereby normalised and is expressed as log copies of the virus/103 cells.
Bacterial cultures
Samples were seeded onto blood agar (BA) and chocolate agar (CHO) and incubated at 37 °C (5% CO2). After 24/48 h colonies morphologically compatible with URTIs were identified using MALDI-TOF (12). Depending on the pathogen isolated, a sensitivity study was carried out following the criteria of EUCAST/NCCLSI (13) which involved microdilution in bacterial broth using the commercial automated system MicroScan (Siemens, Erlangen, Germany).
Biochemical markers
In order to detect biochemical parameters blood samples were taken and stored in lithium heparin tubes to avoid coagulation. Plasma levels of PCT and CRP were established using the Cobas 8000 analyser employing, respectively, electrochemiluminescence (ECLIA) and immunoturbidimetry (Roche Diagnostics® S.L.Mannhein, Germany).
For PCT the reference value of 0.5 ng/mL was used, below which probability of infection is considered to be low. For CRP, the value of 1 mg/dL was established as the cut off in the paediatric population.
Hematological markers
Leukocyte count and formula were carried out on blood samples collected in tubes using EDTA K3 as anticoagulant and processed in a Sysmex XN-1000 analyzer (Sysmex Corporation, Kobe, Japan), which uses flow cytometry fluorescence.
Statistical analyses
The programme MedCalc® Sofware Ltd, version 19.2.1, Ostend, Belgium were used for the statistical analyses. The quantatative variables were expressed as means with standard deviation (M ± SD) and the categorical, or discrete, variables as numbers and percentages. To examine relationships between qualatative variables, a Fisher’s exact test was used and the discrimination capacity of the test was determined using ROC curves (14). A value of P<0.05 was assumed for statistical significance.
Results
During the epidemiological monitoring season studied here, a total of 3,441 paediatric samples were processed, of which 2,162 (62.8%) tested positive for virus: 588 (17.1%) for HAdV, 424 (12.3%) for RSV, 315 (9.2%) for IFVA, 305 (8.9%) ETV, 171 (5%) IFVB, 163 (4.7%) HRV, 86 (2.5%) HCoV, 70 (2%) hMTV and 40 (1.2%) PIV. Figure 1 shows the weekly frequency of the most frequently detected viruses.
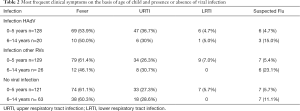
Of the 487 children presenting in the ED who had simultaneous biochemical and haematological tests, 303 (62.2%) had viral respiratory infections. HAdV was detected in 148 of these samples (30.4%), and other respiratory viruses (RVs) in 155 (31.8%), Mixed viral infection was found in 50 cases (10.3%), 34 of which involved HAdV.
Table 2 show the data for this group of children in terms of distribution by age, clinical symptoms at presentation in the ED and type of virus involved (HAdV or other respiratory viruses). Well Table 2 shows that viral infection was present in 257 of the 378 children under 5 (68%) and in 46 (42.2%) of the 109 children aged 6–14 (P<0.0001). It is also worth noting that 63 (57.7%) of the 109 children over 5 had no viral infection.
Full table
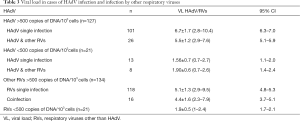
Of the 148 HAdV infected children, viral load was over 500 copies of DNA/103 cells in 127 cases, 101 of which were single infections and 26 coinfections with other respiratory viruses (10 with RSV, 4 with HCoV, 3 with IFBV, 3 with ETV, 2 with IFAV, 2 with PIV, 1 with hMPV and 1 had multiple coinfections with RSV and hMPV. In terms of the 21 children with viral loads of below 500 copies/103 cells, 13 were single infections and 8 coinfections with other respiratory viruses.
In terms of the other non-HAdV respiratory viruses identified, 155 children tested positive (134 of the cases presenting viral loads of over 500 copies/103 cells), with 118 corresponding to single infections (28 IFVA, 26 RSV, 27 ETV, 16 IFVB, 9 HCoV, 5 hMPV, 6 HRV and 1 PIV) and 16 to coinfections. Average viral load values for all groups of viral infection are shown in Table 3.
Full table
Relationship of biomarkers in cases of infection by HAdV, other respiratory viruses and no viral infection
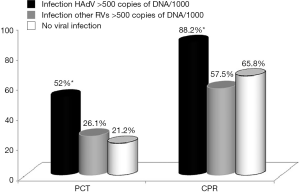
Of the 487 children studied, PCT was found to be above the reference value of 0.5 ng/mL in 146 of the cases (30.0%), 100 of these corresponding to cases where viral load of HAdV or other respiratory viruses was >500 copies of DNA/103 cells, CRP values, on the other hand, were above the cut off of 1 mg/dL in 330 cases (67.7%), 189 of which were in cases with viral load of >500 copies of DNA/103 cells (HAdV or other respiratory viruses) (Figure 2, Table 4).
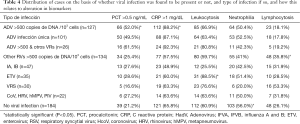
Full table
It can be seen that PCT was altered in 66 HAdV infected children. These corresponded to 50 (49.5%) of the 101 children with single HAdV infection, as well as 16 (61.5%) of those coinfected with HAdV and other respiratory viruses (P>0.05). In addition, 34 (25.4%) of the 134 children with other respiratory viruses, and 39 (21.2%) of the 184 children with no viral infection also had altered PCT (P<0.0001).
The trend was similar, though greater, in the case of CRP, where values were over 1 mg/dL in 112 (88.1%) of the 127 cases where HAdV was present at high viral loads: 88 cases (87.1%) with single HAdV infection and 24 (92.3%) of those with HAdV infection co-occurring with another respiratory virus (P>0.05). A further 77 (57.4%) of the children infected with other respiratory viruses and 121 (65.7%) of those with no viral infection also had elevated CRP (P<0.0001).
In order to evaluate the usefulness of these two biomarkers as diagnostic tests for HAdV, a ROC curve was plotted and the area under the curve (AUC) calculated. This measure indicates the discrimination capacity of each marker in terms of distinguishing whether or not HAdV infection is present (Figure 3).
The AUC for PCT was 0.7 (95% CI: 0.657–0.742) and 0.692 (95% CI: 0.647–0.733) for CRP.
In addition to this, the frequency of infection by HAdV with respect to PCT and CRP values was calculated and the possibility considered of this being related to viral load (Figure 4).
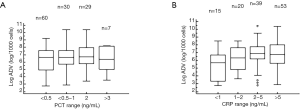
Children with PCT levels in the range 0.5 to 3 ng/mL account for 59 cases (89.4%) of HAdV infection with altered levels of this marker. With respect to CRP, 92 cases (72.4%) had levels above 2 mg/dL and viral load above 6 log, whilst non-pathological levels of CRP were linked with viral loads of 5.2±1.9 (P=0.01).
Haematological markers in cases of infection with HAdV, other respiratory viruses and no viral infection
Of the 487 children studied in this work, 295 (60.7%) had leukocytosis, 240 (49.3%) had neutrophilia and 132 (27.1%) had lymphocytosis. Table 4 shows the results for those children with altered hemograms according to the type of infection always >500 copy/103 cels (single HAdV, HAdV coinfection with other respiratory viruses, other respiratory viruses always >500 copy/103 cels and no viral infection).
Of the 127 children with high viral loads of HAdV, 85 (66.9%) had leukocytosis compared to 80 (59.7%) children with other respiratory virus infections. Of those children with viral infections other than HAdV, significantly more children with ETV manifested this alteration (31 cases, 88.5%, P<0.05) than those with other respiratory viruses. i.e., 12 (25%) with influenza (IFVA or IFVB), 23 (76.6%) with RSV, or the 14 cases (63.6%) where other respiratory viruses were present. What is more, leukocytosis was more common in ETV infected children than in those with no viral infection (112 cases, 60.9%) or those with HAdV.
With respect to neutrophilia, this was experienced by 103 children (56%) without viral infection compared to 64 (50.4%) of those with high viral load of HAdV and 57 (42.5%) with high viral load of other viral infections (P=0.02). Interestingly, the proportion of children with RSV that had neutrophilia (20%) was lower than in other groups with viral infection, including HAdV (P<0.05). Lymphocytosis was found in 45 children (33.5%) with non-HAdV viral infection compared to 23 (18.1%) HAdV infected children (P=0.005).
Bacterial infections
In addition to the identification of viral infection, for 141 of the 487 children studied here a simultaneous bacterial culture was carried out. This resulted in the isolation of bacteria in 16 cases (11.3%) (9 S. pyogenes, 3 H. influenza, 2 S. aureus, 1 S. sciuri, 1 S. disgalactie), whilst in 48 cases (37.3%) HAdV was detected (2 cases of mixed infection with bacteria), in 40 samples (28.3%) other respiratory viruses were identified (9 cases of mixed infection with bacteria) and in 49 cases (34.7%) both the bacterial culture and virus detection proved negative. Of the 16 cases of bacterial infection identified, 11 (68.7%) were mixed infection with virus.
The biomarkers and the hemograms presented by the different groups of patients are shown in Table 5.

Full table
PCT was found to be altered in 27 children (60%) with HAdV infection, in 7 (22.5%) with other respiratory viruses, 3 (18.7%) with bacterial infection and 13 (26.5%) without either viral or bacterial infection (P<0.05).
CRP levels over 1 mg/dL, on the other hand, were found in 40 children (88.9%) with HAdV, 19 (61.3%) of those with other respiratory viruses, 14 (87.5%) with bacterial infection and 36 (73.5%) of those with no viral or bacterial infection (P<0.05).
In terms of haematological markers, it is worth highlighting that leukocytosis was detected in 24 cases (53.3%) of HAdV infection, 14 (87.5%) of bacterial infection and 20 (64.5%) of infection by respiratory viruses other than HAdV (P<0.05). The frequency of neutrophilia ranged from 19 (61.30%) in cases of infection by respiratory viruses other than HAdV to 13 (81.2%) in children with bacterial infection, while lymphocytosis levels were similar across all groups.
Discussion
The accessibility, simplicity and speed of biochemical and hematological markers has made them essential tools in the management of any pathology, including those of an infectious nature (15-17). However, occasionally they may lead to errors if hypotheses are accepted that are not sufficiently corroborated or are not fully applicable to certain groups of patients (18). This study was designed with the aim of clarifying the relationship between viral infection and biochemical as well as haematological markers of inflammation.
Over the period studied, of the 3,441 samples examined from children with respiratory symptoms, 63% had viral infections, the most frequent being HAdV, followed by RSV, IFVA and ETV, despite the fact that the circulation of RSV was very intense during the months of November and December and that of influenza A during the month of January, ADV circulation was more constant without showing peaks that showed RSV or AI.
From all the samples received at the Virology Laboratory, samples from 487 children who had presented at the Paediatric Emergency Department with acute respiratory infection were simultaneously sent for analysis of biomarkers of infection as well as leucocyte formula and count.
The majority of these children were under 6 years of age and the most common symptom they were recorded as experiencing was fever, perhaps because of difficulties in defining the pathology more clearly in such young children. As in the wider study, more than two thirds of the children in this group were found to have viral infection, a rate that is within the range described in the literature, i.e., 55–65% for similar conditions (19). And once again, HAdV was the most frequent viral agent implicated, being found with a normalised viral load of over 5 log (95% CI: 6.1–6.7) in 26.1% of this subgroup. In 80% of these cases, HAdV was found as a single infection, and its frequency was far greater than that of other respiratory viruses, suggesting that HAdV can result in clinical pictures that may necessitate further tests in order to correctly identify the pathological process involved.
In terms of clinical symptoms, HAdV was principally detected in children presenting with fever and pharyngitis-tonsillitis, rather unspecific symptoms with a number of possible, and varied, aetiologies. This could account for the Paediatric Emergency Department requesting tests for inflammation markers (PCT and CRP) as well as haematological markers, which would initially lead to bacterial infection being considered as the cause, in line with publications by other authors (20,21).
However, the analysis reveals that in more than half the cases of HAdV infection with high viral load, levels of PCT were altered, although it should be noted that in 90% of these cases PCT levels did not exceed 1.2 ng/mL. No statistically significant relationship could, however, be established between normalised viral load and PCT values.
With respect to CRP, presence of HAdV was found to lead to alterations in 9 out of every 10 cases. Other respiratory viruses also demonstrated this effect, but not to the same extent. These results suggest that higher levels of CRP correspond to more active viral replication.
Thus it can be seen that there is a relationship between HAdV infection and altered levels of the biochemical markers PCT and CRP, and that this is not found with respect to other respiratory viruses. In addition, the ROC curve analyses carried out revealed that both PCT and CRP tests have an acceptable discrimination capacity to distinguish whether HAdV infection is present or not.
In order to confirm the link found between HAdV infection and the two biomarkers (PCT and CRP), samples were analysed which incorporated factors that might have an influence on them, i.e., bacterial infection. Data from 141 children was analysed with the samples being subjected to not only studies of the viruses involved and the biochemical markers, but also a full microbiological screening in order to establish the aetiological agent responsible for the respiratory infection. Bacteria were only isolated from 16 children (11.3%), a lower rate than those infected by HAdV (33.3%) or even by other respiratory viruses (28.3%). Analyses of these data once again showed that HAdV infection alone was sufficient to alter PCT level, without any concomitant bacterial infection, and confirmed that this relationship was not found with respect to other respiratory viruses. The same association was observed with CRP levels above 1 mg/dL, which were found to be similar in children with bacterial infection to those with HAdV infection, and, in addition, the levels in these two groups were significantly higher than in children infected by other respiratory viruses.
There is currently some doubt that specific levels of these biomarkers is sufficient to identify the type of infection involved in a pathology, and that the kinetics of the pathogen themselves needs to be taken into account in order to reach more reliable conclusions. In this sense, certain authors have described the gradual reduction in PCT in processes such as sepsis or chronic obstructive pulmonary disease (COPD) exacerbation with bacterial origins (22). While the results presented in the current work need to be corroborated, these findings point to the value of using direct microbiological markers, which are increasingly fast and more sensitive, to provide the most appropriate management of patients in terms of complementary tests and avoiding unnecessary treatments.
With respect to the haematological parameters, neither leucocytosis, neutrophilia nor lymphocytosis was found to be a reliable marker for HAdV infection. It should, however, be noted that HAdV tended to be associated with neutrophilia, although, as would be expected, not to such a great extent as in bacterial infections (23,24). However, when the data for respiratory viruses other than HAdV are broken down, children with flu (IFVA and IFVB) had lower levels of leukocytosis than those infected with other viruses, including HAdV or those with no viral infection, while children with ETV infection had the highest levels of leukocytosis. Neutrophilia, on the other hand, was less common in children infected with RSV than in any other group infected with viruses, including HAdV, while lymphocytosis was most common in children infected by non-HAdV viruses, particularly RSV.
Although it was not possible in this work, it would be interesting to know the genotype of the HAdV detected since it has been found that certain genotypes are related to serious respiratory infections, similar to clinical pictures of sepsis that may prove fatal, and that such genotypes might behave in a similar way to bacteria in terms of their relationship with the biomarkers studied here (25).
In summary, HAdV frequently causes respiratory infections in children which are similar to those of bacterial origin, not only clinically, but also in terms of how they alter classic markers for such infections such as PCT and CPR. As such, with respiratory infections of this type it is advisable to discount a viral origin before doing further tests. Distinguishing HAdV infection through means other than the markers discussed would avoid the need for supplementary tests and avoid the unnecessary use of antibiotic.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: We present the following article in accordance with the MDAR checklist. Available at http://dx.doi.org/10.21037/tp-20-333
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-333
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-333). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and has been approved by Hospital Universitario Central de Asturias Ethical Committee (nº 2020.383). Our study is based on the use of remnants of samples used for routine diagnosis. The retrospective collection of the informed consent belonging to the collection of samples stored by the Mycrobiology Service of HUCA was complicated, so the Ethical Committee was asked to exempt informed.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- . ED S. Epidemiology of acute respiratory infection. Sem Pediatric Infect Disease 1998;9:31-6. [Crossref]
- Campbell H. Acute respiratory infection: a global challenge. Arch Dis Child 1995;73:281-3. [Crossref] [PubMed]
- Scott MK, Chommanard C, Lu X, et al. Human Adenovirus Associated with Severe Respiratory Infection, Oregon, USA, 2013-2014. Emerg Infect Dis 2016;22:1044-51. [Crossref] [PubMed]
- Chehadeh W, Al-Adwani A, John SE, et al. Adenovirus types associated with severe respiratory diseases: A retrospective 4-year study in Kuwait. J Med Virol 2018;90:1033-9. [Crossref] [PubMed]
- Walls T, Shankar AG, Shingadia D. Adenovirus: an increasingly important pathogen in paediatric bone marrow transplant patients. Lancet Infect Dis 2003;3:79-86. [Crossref] [PubMed]
- Andreola B, Bressan S, Callegaro S, et al. Procalcitonin and C-reactive protein as diagnostic markers of severe bacterial infections in febrile infants and children in the emergency department. Pediatr Infect Dis J 2007;26:672-7. [Crossref] [PubMed]
- Schuetz P, Wirz Y, Sager R, et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev 2017;10:CD007498 [Crossref] [PubMed]
- Huang DT, Yealy DM, Filbin MR, et al. Procalcitonin-Guided Use of Antibiotics for Lower Respiratory Tract Infection. N Engl J Med 2018;379:236-49. [Crossref] [PubMed]
- Betakova T, Kostrabova A, Lachova V, et al. Cytokines Induced During Influenza Virus Infection. Curr Pharm Des 2017;23:2616-22. [Crossref] [PubMed]
- Bhuiyan MU, Blyth CC, West R, et al. Combination of clinical symptoms and blood biomarkers can improve discrimination between bacterial or viral community-acquired pneumonia in children. BMC Pulm Med 2019;19:71. [Crossref] [PubMed]
- Alvarez-Arguelles ME, de Ona-Navarro M, Rojo-Alba S, et al. Quantification of human papilloma virus (HPV) DNA using the Cobas 4800 system in women with and without pathological alterations attributable to the virus. J Virol Methods 2015;222:95-102. [Crossref] [PubMed]
- Clark AE, Kaleta EJ, Arora A, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 2013;26:547-603. [Crossref] [PubMed]
- Leclercq R, Canton R, Brown DF, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 2013;19:141-60. [Crossref] [PubMed]
- Cerda J, Cifuentes L. Using ROC curves in clinical investigation: theoretical and practical issues. Rev Chilena Infectol 2012;29:138-41. [Crossref] [PubMed]
- O'Riordan F, Shiely F, Byrne S, et al. An investigation of the effects of procalcitonin testing on antimicrobial prescribing in respiratory tract infections in an Irish university hospital setting: a feasibility study. J Antimicrob Chemother 2019;74:3352-61. [Crossref] [PubMed]
- Branche AR, Walsh EE, Vargas R, et al. Serum Procalcitonin Measurement and Viral Testing to Guide Antibiotic Use for Respiratory Infections in Hospitalized Adults: A Randomized Controlled Trial. J Infect Dis 2015;212:1692-700. [Crossref] [PubMed]
- Chang CH, Tsao KC, Hu HC, et al. Procalcitonin and C-reactive protein cannot differentiate bacterial or viral infection in COPD exacerbation requiring emergency department visits. Int J Chron Obstruct Pulmon Dis 2015;10:767-74. [Crossref] [PubMed]
- Lacoma A, Prat C, Andreo F, et al. Value of procalcitonin, C-reactive protein, and neopterin in exacerbations of chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 2011;6:157-69. [PubMed]
- Kumar P, Medigeshi GR, Mishra VS, et al. Etiology of Acute Respiratory Infections in Infants: A Prospective Birth Cohort Study. Pediatr Infect Dis J 2017;36:25-30. [Crossref] [PubMed]
- Mahajan P, Grzybowski M, Chen X, et al. Procalcitonin as a marker of serious bacterial infections in febrile children younger than 3 years old. Acad Emerg Med 2014;21:171-9. [Crossref] [PubMed]
- Lepe Jiménez JA, Vázquez Florido A, Ramos de Mora M, et al. Utility of procalcitonin, C-reactive protein and leukocyte count in the detection of meningococcal disease during an epidemic cluster. An Pediatr (Barc) 2005;62:328-32. [PubMed]
- Billeter A, Turina M, Seifert B, et al. Early serum procalcitonin, interleukin-6, and 24-hour lactate clearance: useful indicators of septic infections in severely traumatized patients. World J Surg 2009;33:558-66. [Crossref] [PubMed]
- Honda T, Uehara T, Matsumoto G, et al. Neutrophil left shift and white blood cell count as markers of bacterial infection. Clin Chim Acta 2016;457:46-53. [Crossref] [PubMed]
- West DC, Andrada E, Azari R, et al. Predictors of bacteremia in febrile children with sickle cell disease. J Pediatr Hematol Oncol 2002;24:279-83. [Crossref] [PubMed]
- Suberviola B, Castellanos-Ortega A, González-Castro A, et al. Prognostic value of procalcitonin, C-reactive protein and leukocytes in septic shock. Med Intensiva 2012;36:177-84. [Crossref] [PubMed]




