
PEX26 gene genotype-phenotype correlation in neonates with Zellweger syndrome
Introduction
Zellweger spectrum disorder (ZSD) is a phenotypic continuum including Zellweger syndrome (ZS; OMIM #214100), neonatal adrenoleukodystrophy (NALD; OMIM #202370) and infantile Refsum disease (IRD, OMIM #266510). ZS is the most severe form of ZSD and is characterized by craniofacial dysmorphism and severe neurological abnormalities (1,2). Life expectancy in infants with ZS is severely limited, with rarely a one-year survival (3). Patients with ZS have severe hypotonia, difficulty feeding, hepatomegaly, ocular disorders, and seizures. In addition, this phenotype is characterized by craniofacial abnormalities, including widely fontanelle, cranial sutures, prominent forehead, and low-set ears (4).
ZSD is an autosomal recessive inherited disorder caused by a genetic defect (called PEX) involved in peroxisomal biosynthesis. Up to now, there have been 14 ZSD-PEX genes associated with ZSD (5). These PEX genes encode proteins required for peroxisomal biogenesis, including membrane biogenesis, division, and include the mechanism of peroxisomal matrix introduction (6,7). Among all ZSD patients, PEX1 mutation was the most common cause, accounting for 58%. Nevertheless, PEX26 mutations account for only 3% of total ZSD (8). Therefore, the phenotype of ZS caused by PEX26 gene mutation is wide and dispersed.
In this study, we reported a neonatal ZS case with a novel homozygous mutation in PEX26 gene. We also searched and retrospectively analyzed PEX26 mutations in neonatal cases diagnosed with ZS from the database. This research enriches the clinical features and evaluates the phenotypic and genotypic associations induced by ZSD with PEX26 mutations.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-103).
Methods
Case presentation
We investigated the case of a near-term infant, female, born at 36+6 weeks gestation, weighing 2,230 g. The infant was transferred to the Neonatal Intensive Care Unit (NICU) of the Children’s Hospital of Fudan University at 3 hours after birth, due to polypnea and birth asphyxia. Mom had normal prenatal care. Fetal ultrasonography at 28 weeks revealed bilateral cerebral ventriculomegaly, and fetal magnetic resonance imaging (MRI) manifested enlargement of posterior horn of right ventricle. Her parents were healthy and non-consanguineous. The infant was born with hypotonia and had no spontaneous respiration, required intubation at birth. Her Apgar scores were 4 at 1 minute, 4 at 5 minutes and 8 at 10 minutes respectively. Physical examination on admission revealed large anterior and posterior fontanel with prominent forehead and widely separated sutures, low-set ears, systemic hypotonia, minimal spontaneous movement, and transverse palmar creases on the left hand.
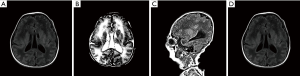
Initial laboratory workup demonstrated hyperbilirubinemia (327.6 µmol/L) and elevated liver enzymes (aspartate aminotransferase, 245.3 IU/L; alanine aminotransferase, 143.3 IU/L), suggesting hepatic dysfunction. Urine organic acids revealed obvious elevated 4-hydroxyphenyllactic acid. Very long chain fatty acid of plasma analysis showed normal C22 and C24, but dominant elevated C26 (>11.69 nmol/L, normal <1.3 nmol/L), C24/C22 (2.32, normal ≤1.39), C26/C22 (>0.649, normal ≤0.023). The chest X-ray showed right pneumothorax. Abdominal ultrasound showed indistinct structure of bilateral kidneys. Head ultrasound indicated bilateral cerebral ventriculomegaly, which confirmed by fetal cerebral MRI (Figure 1). A bleeding spot was found on the right fundus by ophthalmic examination. During hospitalization, she was initially treated with mechanical ventilated and ursodeoxycholic acid. She was discharged home with low flow nasal prone oxygen and fed by nasogastric tube due to hypotonia and weak uncoordinated suck reflex.
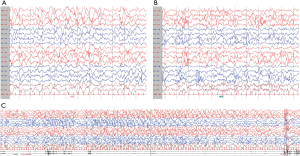
The infant developed an epileptic seizure when she was eight months old and was brought to our hospital for treatment. Video-electroencephalogram (VEEG) showed hypsarrhythmia during both wakefulness and sleep, especially in the posterior area. Moreover, focal seizures were detected during sleep (Figure 2). With palliative care, she died at 9 months of age from dyspnea and cardiopulmonary arrest.
Whole exome sequencing (WES)
A trio WES was performed by WuXi NextCODE Genomics, Shanghai, China (CLIA Lab ID: 99D2064856) using a previous described protocol. Briefly, exome capture was performed using the Agilent SureSelect Human All Exon V5, Illumina TruSeq Rapid PE Cluster, and SBS kits (Agilent Technologies, Santa Clara, CA, USA). WES was performed on the Illumina HiSeq 2000/2500 platform. Reads were aligned to the human genome reference sequence (GRCh37/hg19 build of UCSC Genome Browser; http://genome.ucsc.edu) with the Burrows-Wheeler Aligner v.0.6.2. Duplicate paired-end reads were marked with Picard v.1.55 (https://broadinstitute.github.io/picard/). The Genome Analysis Toolkit v.2.3–9 was used for base quality score recalibration, indel realignment, and variant discovery. Variants were annotated using a pipeline developed in-house and filtered in the Exome Variant Server, gnomAD, Exome Aggregation Consortium, or the dbSNP databases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Children’s Hospital of Fudan University (IRB No.2021-237). The parents of the patient provided written informed consent.
Literature review and data analysis
A comprehensive literature search was performed to identify the relevant researches and case reports in PubMed, the Web of Science and EMBASE from inception to August 1, 2020. We used the following search strategy “((PEX26) OR (PEX26 gene)) AND ((Zellweger syndrome) OR (ZS)OR (ZSD) OR (Peroxisome biogenesis disorder) OR (Zellweger spectrum disorder))”. In the Chinese database, literature researches were conducted by using the “PEX26”, “Zellweger syndrome”, and “Peroxisome biogenesis disorder” as keywords from the inception to August 1, 2020, and no relevant literatures were found.
Satistical analysis
The genotypes of ZSD with the mutation of PEX26 and their clinical subtypes were compared. Differences in the occurrence of deleterious variants in ZS groups and NALD/IRD groups were analyzed by using Fisher exact test. A two-sided P value was used to test for statistical significance (P<0.05).
Results
Whole exome sequencing
Our case depended on clinical manifestations and laboratory studies for diagnosis. and confirmation is based on the homozygosity variant in the PEX26 gene (NM_017929: exon2: c.34del) that we identified. The variant was validated by Sanger sequencing on a 3500XL instrument, and both parents are heterozygous carriers. We identified this variant as pathogenic variant because the type of this variant is nonsense, which loss of function is a known mechanism of this gene. Moreover, this variant is absent from ExAC, the 1000 Genomes database, and HGMD, ClinVar database.
Clinical characteristic of newborns with ZS caused by PEX26 mutation
A total of 9 articles referring to 31 cases of ZSD with PEX26 pathogenic variants were identified form the electronic database (9-17). However, after careful filtering, only four ZS infants were described in detail in postnatal clinical manifestations. Other babies were mentioned only briefly about the mutation site or were not found to have neonatal presentation. These four infants are from Saudi Arabia (9), Tunisian (10) and America (11,12). Finally, combined with our case, the clinical phenotypes of 5 ZS infants were summarized and shown in Table 1 (male =4, female =1). All (100%, 5/5) infants were characterized by hypotonia, with 80% (4/5) having difficulty feeding, but only 40% (2/5) presenting with seizures. All (100%, 5/5) infants were born with facial deformities and liver dysfunction. 80% (4/5) of the patients were characterized by enlarged fontanelles and prominent forehead. 60% (3/5) of patients had wide cranial sutures and low-set ears. Kidney abnormalities occurred in only 40% of cases (2/5), only one of the patients existed renal cysts. Other systems involved are ventricular dilation, fundus lesions, and skeletal abnormalities (such as bilateral clubfeet and chondrodysplasia). All plasma analyses of very long chain fatty acids showed elevated C26 and/or C24. The prognosis is poor, with three out of five infants failing to survive the first year of life.

Full table
The genotype-phenotype correlation of PEX26 gene
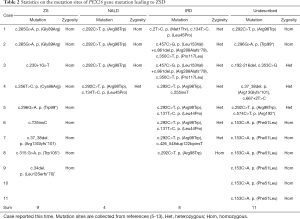
Among the 32 pathogenic variants (our case and the 31 previously reported ones were reviewed), 9 cases were diagnosed as ZS, 4 as NALD, 8 as IRD, and 11 cases were not clinically diagnosed, as shown in Table 2. All showed recessive models, 21 (65.6%) homozygous variants and 11 (34.4%) complex heterozygous variants were identified. It is worth noting that all the mutations in ZS are homozygous, although it contains several types of variation (frameshift, nonsense, missense, and splicing mutations). Therefore, complex heterozygous mutations were found only in NALD and IRD cases, mainly in IRD cases (7/8, 87.5%).
Full table
Currently, 22 disease-causing mutations of ZSD have been reported in the PEX26 gene identified in these 32 patients. Among them, missense variant accounted for 45.5% (10/22), nonsense variant accounted for 22.7% (5/22), frameshift variant accounted for 22.7% (5/22), and splicing variant accounted for 9.1% (2/22). Notably, the missense variant was the most common form of all mutations (42/66, 63.64%) and was identified in 24 patients (3 with ZS, 4 with NALD, 8 with IRD, and 9 with unknown diagnosis). C. 292C>T variant was detected in both NALD and IRD, accounting for the largest proportion (16/66, 24.24%), but it has not been reported in ZS.
Of these 32 patients, 9 were diagnosed with ZS, of which 3 had missense mutations (33.3%, 6/18), and the remaining 6 had harmful mutations (66.7%, 12/18), including nonsense, splices, or frameshift mutations. Another 12 patients were diagnosed with NALD or IRD, with missense mutations and deleterious mutations accounting for 84.6% (22/26) and 15.4% (4/26) of the total mutations, respectively (Table 2). The proportion of harmful mutations in ZS patients was significantly higher than that in NALD and IRD patients (P<0.001) (Table 3).

Full table
Discussion
Considering its pathophysiology, ZSD has been classified into three categories. The first group: ZS, critically sick newborns, most of whom do not survive beyond the first year. Group 2: NALD, infantile, usually has adrenal cortex atrophy, and most children die before the age of three to five years. Group 3: IRD, mild. Most of those affected suffer from ataxia, and many of them can go into adolescence (3,18,19). The differences between the three diseases were due to the severity of the clinical phenotype and the mutation of the genotype. Recently, Heimler syndrome, a very mild and unique phenotype associated with defects in the PEX gene, has been reported. Heimler syndrome is considered to be part of the ZSD continuum (20,21), but is not generally recognized.
ZS is a multisystem disease that is apparent at birth. Findings include severe characteristic craniofacial deformities, hypotonia, and seizures. Hepatomegaly and renal cortical cyst accounted for 80% and 70% respectively (3). Eye abnormalities such as cataracts and sensorineural hearing loss are typical but not always recognizable when first presented (22). Our literature review found that in all ZS infants with PEX26 mutations, hypotonia, facial deformities, abnormal liver function and elevated C26 / C24 were prominent signs. However, the incidence of seizures in the neonatal period (40%) is lower than that of hypotonia and liver disease, which may be related to atypical at the neonatal period or too early death to present. Meanwhile, PEX1 was the most involved gene, accounting for nearly 70% of patients with ZSD. Preuss et al. found that renal cysts are typical of PEX1 mutated infants (23), which is different from our results (only one of five renal cysts exists). With respect to bone signs, two-fifths of the newborns in our study showed bilateral clubfeet, which has not been reported in ZS infants with PEX1 mutation. Brain MRI or ultrasound revealed polymicrogyria and ventriculomegaly in 80% of ZS infants with PEX26 mutations. These brain disorders may contribute to the diagnosis of the disease.
PEX26 encodes a 34-kDa peroxisomal membrane protein of 305 amino acids, that assists in the localization of PEX1-PEX6 complexes to the peroxisomal membrane (6,15,24). The C-terminal part of PEX26 is exposed to the matrix side with one transmembrane domain (aa252-269) located in the peroxisomal membrane, while the N-terminal part is cytosol-exposed and vital in its biological activity. The PEX6 binding domain located in the N-terminal region of the protein corresponds to the amino acid 29-174 (14,17,25), which is essential for binding to PEX6. The PEX1-PEX6 AAA ATPase complex was recruited and anchored to the peroxisomal membrane (26,27). However, Tamura et al. believed that PEX26 residues amino acids13–48 was sufficient to bind to PEX6 (28).
The strongly represented mutation in ZSD with PEX26 mutations is the missense mutation c.292C>T, which changes the arginine located at amino acid position 98 into a tryptophane (p. Arg98Trp). This missense mutation affects the stable expression of PEX26. The PEX6 binding domain is destroyed and the ability to participate in the interaction between PEX6 and PEX1 is reduced (27,28). However, the effect of recurrent c.292C> T mutation was relatively minor. It has been reported not only in NALD but also in IRD. The same mutation suggests that different clinical phenotypes, which may be related to the temperature sensitivity of PEX26 cell lines (14). Since this mutation has not yet been identified in ZS, we infer that the missense mutation c.292C>T causes residual protein levels and function, and thus exhibits a relatively mild clinical phenotype.
Mutations of PEX26 are distributed across the entire gene, but 70% (14/20) of the deleterious mutations of ZS are located in the N-terminal part (Figure 3), including nonsense, frameshift and splicing mutations, that theoretically causes a truncation of functional domain and/or large domain deletion in peroxins, resulting in dysfunction. The closer the mutation site is to the N terminal, the greater the damage to the structure and homeostasis of peroxisomes. ZS patient-derived PEX26 mutants seriously impaired peroxisomal matrix protein import, which progressively influence the energy GTP generated and provided for the fission of peroxisomes (29). Thus, these mutations are total or almost total loss of protein levels and function, and are most severe in patients with ZS. In contrast, missense variants that retain partial function have less effect on peroxisome assembly (30,31). Patients with NALD or IRD, mainly composed of missense mutations, showed moderate or mild clinical phenotypes. The severity of the mutation depends on the effect on the function of PEX26. Our result showed that the proportion of deleterious mutation in ZS is significantly higher than NALD/IRD (P<0.05). Thus, this may explain why the ZS phenotype is more severe than other phenotypes. The type of PEX26 mutation can help predict the severity of individual cases.

In conclusion, we reported a ZS neonate developed facial deformities, hypotonia, feeding difficulties, and seizures. The new pathogenic mutation c. 34del was found in the PEX26 gene. We also summarized the manifestations of reported ZS neonates with PEX26 mutation, including characteristic craniofacial dysmorphism, hypotonia, cerebral dysfunctions (cerebral dysfunctions), liver dysfunction. Elevated C26 and/or C24 in VLCFA is usually shown in blood plasma analysis. However, seizures and renal cysts are less obvious for neonatal presenting in newborns with PEX26 mutation. In addition, the c.292C>T mutation occurred at a relatively high frequency in ZSD with the PEX26 mutation, which resulted in a relatively mild clinical phenotype. Finally, we compared the occurrence of deleterious variants, associated with a disease severity, between ZS groups and NALD/IRD groups, ZS accounting for the significant higher percentage and therefore demonstrated the most severe clinical manifestation.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-103
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tp-21-103
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-103). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Children’s Hospital of Fudan University (IRB No.2021-237). The parents of the patients provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Braverman NE, Raymond GV, Rizzo WB, et al. Peroxisome biogenesis disorders in the Zellweger spectrum: An overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol Genet Metab 2016;117:313-21. [Crossref] [PubMed]
- Honsho M, Okumoto K, Tamura S, et al. Peroxisome Biogenesis Disorders. Adv Exp Med Biol 2020;1299:45-54. [Crossref] [PubMed]
- Cheillan D. Zellweger Syndrome Disorders: From Severe Neonatal Disease to Atypical Adult Presentation. Adv Exp Med Biol 2020;1299:71-80. [Crossref] [PubMed]
- Wanders RJA. Metabolic and molecular basis of peroxisomal disorders: A review. Am J Med Genet 2004;126A:355-75. [Crossref] [PubMed]
- Fujiki Y. Peroxisome biogenesis and human peroxisome-deficiency disorders. Proc Jpn Acad Ser B Phys Biol Sci 2016;92:463-77. [Crossref] [PubMed]
- Waterham HR, Ebberink MS. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta 2012;1822:1430-41. [Crossref] [PubMed]
- Waterham HR, Ferdinandusse S, Wanders RJ. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta 2016;1863:922-33. [Crossref] [PubMed]
- Ebberink MS, Mooijer PA, Gootjes J, et al. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum Mutat 2011;32:59-69. [Crossref] [PubMed]
- Al-Sayed M, Al-Hassan S, Rashed M, et al. Preimplantation genetic diagnosis for Zellweger syndrome. Fertil Steril 2007;87:1468.e1-3. [Crossref] [PubMed]
- Ben Turkia H, Yangui M, Azzouz H, et al. A novel mutation in PEX 26 gene in Zellweger syndrome: a case report. Tunis Med 2011;89:288-91. [PubMed]
- Stowe RC, Agarwal S. Novel PEX26 Mutation Causing Zellweger Syndrome Presenting as Feeding Intolerance and Hypotonia. Pediatr Neurol 2017;75:96-7. [Crossref] [PubMed]
- Rife E, Dumbar AE, Nelson SL, et al. Stippled Chondral Calcifications of the Patella in Zellweger Syndrome. J Pediatr 2018;192:265. [Crossref] [PubMed]
- Tanaka AJ, Okumoto K, Tamura S, et al. A newly identified mutation in the PEX26 gene is associated with a milder form of Zellweger spectrum disorder. Cold Spring Harb Mol Case Stud 2019;5:a003483 [Crossref] [PubMed]
- Matsumoto N, Atsumoto N, Tamura S, et al. Mutations in Novel Peroxin Gene PEX26 That Cause Peroxisome-Biogenesis Disorders of Complementation Group 8 Provide a Genotype-Phenotype Correlation. Am J Hum Genet 2003;73:233-46. [Crossref] [PubMed]
- Furuki S, Tamura S, Matsumoto N, et al. Mutations in the peroxin Pex26p responsible for peroxisome biogenesis disorders of complementation group 8 impair its stability, peroxisomal localization, and interaction with the Pex1p x Pex6p complex. J Biol Chem 2006;281:1317-23. [Crossref] [PubMed]
- Steinberg S, Chen L, Wei L, et al. The PEX Gene Screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Mol Genet Metab 2004;83:252-63. [Crossref] [PubMed]
- Weller S, Cajigas I, Morrell J, et al. Alternative Splicing Suggests Extended Function of PEX26 in Peroxisome Biogenesis. Am J Hum Genet 2005;76:987-1007. [Crossref] [PubMed]
- Percy AK, Rutledge SL. Adrenoleukodystrophy and related disorders. Ment Retard Dev Disabil Res Rev 2001;7:179. [Crossref] [PubMed]
- Bousfiha A, Bakhchane A, Charoute H, et al. A novel PEX1 mutation in a Moroccan family with Zellweger spectrum disorders. Hum Genome Var 2017;4:17009. [Crossref] [PubMed]
- Yik WY, Steinberg SJ, Moser AB, et al. Identification of Novel Mutations and Sequence Variation in the Zellweger Syndrome Spectrum of Peroxisome Biogenesis Disorders. Hum Mutat 2009;30:E467-80. [Crossref] [PubMed]
- Ratbi I, Falkenberg KD, Sommen M, et al. Heimler Syndrome Is Caused by Hypomorphic Mutations in the Peroxisome-Biogenesis Genes PEX1 and PEX6. Am J Hum Genet 2015;97:535-45. [Crossref] [PubMed]
- Klouwer FCC, Berendse K, Ferdinandusse S, et al. Zellweger spectrum disorders: clinical overview and management approach. Orphanet J Rare Dis 2015;10:151. [Crossref] [PubMed]
- Preuss N, Brosius U, Biermanns M, et al. PEX1 mutations in complementation group l of Zellweger spectrum patients correlate with severity of disease. Pediatr Res 2002;51:706-14. [Crossref] [PubMed]
- Nazarko TY. Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders. Autophagy 2017;13:991-4. [Crossref] [PubMed]
- Zhang R, Chen L, Jiralerspong S, et al. Recovery of PEX1-Gly843Asp peroxisome dysfunction by small-molecule compounds. Proc Natl Acad Sci U S A 2010;107:5569-74. [Crossref] [PubMed]
- Guder P, Lotz-Havla AS, Woid M, et al. Isoform-specific domain organization determines conformation and function of the peroxisomal biogenesis factor PEX26. Biochim Biophys Acta Mol Cell Res 2019;1866:518-31. [Crossref] [PubMed]
- Matsumoto N, Tamura S, Fujiki Y. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 2003;454-60. [Crossref] [PubMed]
- Tamura S, Matsumoto N, Takeba R, et al. AAA peroxins and their recruiter Pex26p modulate the interactions of peroxins involved in peroxisomal protein import. J Biol Chem 2014;289:24336-46. [Crossref] [PubMed]
- Fujiki Y, Abe Y, Imoto Y, et al. Recent insights into peroxisome biogenesis and associated diseases. J Cell Sci 2020;133:s236943 [Crossref] [PubMed]
- Weller S, Gould SJ, Valle D. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet 2003;4:165-211. [Crossref] [PubMed]
- Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 2006;75:295-332. [Crossref] [PubMed]


