
Bioinformatics analysis reveals the roles of cytoskeleton protein transgelin in occurrence and development of proteinuria
Introduction
Proteinuria, one of the principle signs of chronic kidney disease, is a major risk factor for the progression of renal diseases to end-stage renal failure. Proteinuria is caused by the abnormal transglomerular passage of proteins due to enhanced permeability of the glomerular capillary wall resulting in disabled reabsorption by epithelial cells of the proximal tubuli. Therefore, proteinuria is not only a hallmark of many glomerular diseases, but also a sensitive marker for the progression of kidney disease to renal failure and an independent risk factor for cardiovascular morbidity and mortality (1,2). Proteinuria is closely associated with structural and functional defects of podocytes (3,4). Podocytes are highly specialized and terminally differentiated, featuring a complex cellular morphology with an extensive network of protrusions that extend from the thick primary processes into numerous fine foot processes. Foot processes that are supported by microtubular and actin cytoskeletons (5,6) not only participate in formation of the filtration barrier, but are also essential for increasing surface area, thereby mediating efficient attachment to the glomerular basement membrane (7,8). The effacement of podocyte foot processes is demonstrated to be a well-known occurrence in many glomerulonephritides with proteinuria. Proteinuria occurs when the structure of podocytes is destroyed by disruption of the slit-diaphragm and effacement of foot processes but reverses as the foot processes are recovered (3). The cytoskeleton of podocyte foot processes is formed by F-actin (F-actin)-containing microfilaments, and provides the molecular basis for regulating podocyte shape, structure, stability, slit diaphragm insertion, adhesion, plasticity, and dynamic response to environmental stimuli (9). With the development of the “omics” technologies, more than 50 mutated genes that negatively affect the structural integrity and function of the glomerular filtration barrier, thus contributing to the development of proteinuria, have been identified (2,7,10). Mutated genes of interest include genes coding for slit diaphragm, actin cytoskeleton, mitochondria, glomerular basement membrane (GBM)-related proteins, nuclear transcription factors and proteins of the proximal tubule protein absorption pathways (10), among which actin cytoskeleton-related genes (11-14) account for a significant proportion (7) to cause profound changes in the podocyte actin cytoskeleton. A subset of genetic mutations has been proven to cause nephrotic syndrome that is responsive to immunosuppressive therapeutic schemes. Therefore, interventions targeting pathways that regulate the actin cytoskeleton play an important role in the treatment of proteinuric kidney diseases.
Our previous study identified key differentially expressed genes (DEGs) and cytoskeleton proteins (15) that are involved in development of proteinuria and dynamics of foot processes using GeneChip analysis. Transgelin (TAGLN) is a 22 kDa microfilament-binding protein encoded by a transforming growth factor beta (TGFβ)-inducible gene (16,17). TAGLN has been shown to participate in cytoskeletal organization and contractile regulation by interacting with actin (18). The amino-terminal of TAGLN contains a single calponin homology (CH) domain that is known to play critical roles in linking signaling molecules to the actin cytoskeleton (18). Skeletal protein TAGLN is significantly altered with changes in podocyte foot processes, leading to development of proteinuria in rats with puromycin aminonucleoside (PAN) nephropathy (19-21). TAGLN was observed to be abnormally expressed and distributed in human kidney tissues from minimal-change nephrotic syndrome (MCNS), focal segmental glomerulosclerosis (FSGS) and membranous nephropathy (MN). In the case of podocyte injury, transgelin expression was significantly upregulated in children with MCNS, FSGS and MN, and rats with PAN, and PAN-treated podocytes (15,18,22). Expression of TAGLN was induced in glomerular visceral and parietal epithelial cells in a rat model of anti-glomerular basement membrane (GBM) crescentic glomerulonephritis (22). Recently, Hauser et al. reported significantly elevated expression of transgelin in podocytes of passive Heymann’s nephritis (PHN, experimental membranous nephropathy) (23). The expression of TAGLN was upregulated during podocyte injury induced by AngII, adriamycin and D-glucose (24). However, little is known regarding molecular upstream and downstream regulatory mechanisms underlying the expression and distribution of TAGLN during the development of proteinuria.
The objectives of the present study are to analyze the expression and distribution of skeletal protein transgelin induced by various injury factors and its effects on the cytoskeleton architecture and proteinuria, and elucidate molecular mechanisms of key signaling pathways that regulate TAGLN expression. The findings of the present study further revealed the regulatory role of cytoskeletal protein transgelin in the dynamic changes of podocyte structure and function, and identified the regulatory molecules governing the podocyte actin cytoskeleton, as well as potential interventional targets for proteinuria.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-83).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data collection, information and processing
The National Center for Biotechnology Information (NCBI) GEO database (http://www.ncbi.nlmNih.gov/geo) is a public genomics data repository that contains gene expression datasets and original series and platform records. Microarray datasets GSE108109, GSE99340, GSE125779, GSE108112, GSE121211, GSE108629, GSE124622, GSE35488 and GSE75693 were download from the GEO database, and normalized by Log standardization in R language (version 3.5.0; http://www.r-project.org/) into expression matrices for subsequent analysis.
GSE108109 is from the GPL19983 platform [HuGene-2_1-st] Affymetrix Human Gene 2.1 ST Array, HuGene21st_Hs_ENTREZG_19.0.0] with molecular targets and total RNA expression profile in the glomerular transcriptome from 6 healthy samples (living donor) and 111 cases. GSE125779 and GSE121211 is from the platform GPL17586 [HTA-2_0] Affymetrix Human Transcriptome Array 2.0 [transcript (gene) version]. GSE125779 contains transcriptome comparison of tubulointerstitial tissues from kidneys with focal segmental glomerulosclerosis (FSGS) and tubulointerstitial tissues from the unaffected portion of surgical nephrectomies, whereas GSE121211 contains transcriptome comparison of tubular tissues from kidneys with 5 FSGS and tubular tissues from the unaffected portion of tumor nephrectomies and 5 controls. GSE108112 is from the platform GPL19983 [HuGene-2_1-st] Affymetrix Human Gene 2.1 ST Array [HuGene21st_Hs_ENTREZG_19.0.0] with shared molecular targets in the tubulointerstitial transcriptome from patients with nephrotic syndrome (NS) and antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV). GSE108629 is from the platform GPL10787 Agilent-028005 SurePrint G3 Mouse GE 8x60K Microarray (Probe Name version) with gene expression profiles of normal and injured podocytes induced by immunotoxin. GSE124622 is from platform GPL16791 Illumina HiSeq 2500 (Homo sapiens) with NGS-derived transcriptome profiling (RNA-seq) in control (untreated) and injured (Puromycin aminonucleoside (PAN) and Adriamycin) podocyte. GSE75693 is from the platform GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array with urine proteomics and gene expression data from kidney biopsies from kidney transplant patients with and without acute rejection, chronic allograft nephropathy and BK virus nephritis. GSE99340 is from the GPL19109 platforms [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [CDF: Brainarray HGU133Plus2_Hs_ENTREZG_v18] and GPL19184 [HG-U133A] Affymetrix Human Genome U133A Array [Custom Brainarray v18 ENTREZG CDF], containing the total RNA expression profiles of podocyte AB81 from 3 hypoxic samples and 3 controls. GSE35488 is from the GPL19109 platforms GPL14663 with expression data from human with IgA nephropathy (IgAN) [HG-U133A_ENTREZG_10].
Identification and series test of cluster (STC) analysis of DEGs
The DEGs for nephropathy between 10 comparisons of 5 different nephropathy groups [NS, ANCA, MCD, membranous glomerulonephritis (MGN), FSGS] and control in GSE108109 were identified with the Limma package (version 3.34.8) in the R language and applied for overlap analysis. Comparison groups included FSGS vs. ANCA, FSGS vs. control, FSGS vs. MGN, FSGS vs. MCD, ANCA vs. control, ANCA vs. MGN, ANCA vs. MCD, control vs. MGN, control vs. MCD and MGN vs. MCD. P values were adjusted by the Benjamini-Hochberg (BH) false discovery rate (FDR) method, and adjusted P value (adj.P) <0.05 and |log FC| >1 were used as the cutoff criteria. The overlapped DEGs among 10 comparisons in GSE108109 were selected for STC analysis to identify the most representative gene clusters corresponding to the trend of transgelin expression in nephropathy. STC is a type of hierarchical cluster method to analyze a set of unique model expression tendencies of the overlapped DEGs in accordance with signal density change tendency of genes under different situations by Short Time-series Expression Miner (STEM) (25) (http://www.cs.cmu.edu/~jernst/stem). In addition, DEGs for FSGS vs. control in GES125779 were identified using adjusted P value (adj.P) <0.05 and |log FC| >1 as the cutoff criteria.
The DEGs for normal oxygen vs. hypoxia injured podocytes in GSE99340 and GSE35488, and Adriamycin vs. control, and PAN vs. control in GSE124622, were characterized with the Limma package (version 3.34.8) in the R language. P values were adjusted by the Benjamini-Hochberg (BH) false discovery rate (FDR) method, and adjusted P value (adj.P) <0.05 and |log FC| >2 were used as the cutoff criteria.
Gene Ontology (GO) annotation and pathway enrichment analysis of DEGs
Associations of transgelin with the common genes between DEGs from 10 comparisons in GSE108109 and DEGs in GSE99340 were estimated to determine significant DEGs for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. KEGG pathway analysis was utilized with the Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) database (26) to identify functional and metabolic pathways significantly associated with TAGLN, and any P value <0.05 was rated to be statistically significant. Pathways were constructed based on the interactions in the KEGG database. The coexpression network and gene signal expression networks were developed and analyzed, and the bubble chart was generated using the “ggplot2” package (version 3.1.0).
The interactive associations among the overlapped DEGs between DEGs from 10 comparisons in GSE108109 and DEGs in GSE99340, as well as the DEGs in GSE35488 were evaluated by the Search Tool for the Retrieval of Interacting Genes (STRING) (https://string-db.org/) (27), with a confidence score of >0.4 as the cut-off criterion. STRING is designed to integrate and rank associations from high-throughput experimental data, from the mining of databases and literature, and from predictions based on genomic context analysis across organisms. STRING currently features a library of over 9,643,763 proteins from 2031 organisms (28). Cytoscape software (version 3.5.1; www.cytoscape.org) (29) was applied to construct the PPI network. The hub genes in the network were detected to characterize key elements in TAGLN, using cytoHubba (30) (version 0.1) in Cytoscape.
The overlapped DEGs among GSE125799 (FSGS vs. control), GSE108109 (FSGS vs. control, and MCD vs. control) and GSE124622 (Adriamycin vs. control, and PAN vs. control) were responsible for podocyte injury and nephropathy, and applied for GO annotation and KEGG pathway enrichment analysis.
The interactive associations among 2 genes (TAGLN, transient receptor potential cation channels TRPC) and 2 signaling pathways (P13K-Akt and PPAR) were evaluated by STRING. Genes with combined score above 400 in the signaling pathway were selected for interactive association analysis.
Correlation analysis of TAGLN-related Transcription factors (TFs) and immunity
TFs-regulating TAGLN were screened from the transcription factor database Ensembl (https://asia.ensembl.org). The associations of expression levels of common DEGs between GSE99340 and GSE108109 with TFs were estimated to screen significant TAGLN-related TFs with “cor.test” in the R language.
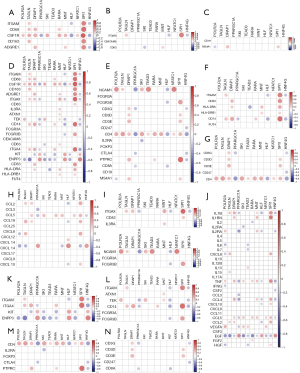
A set of notable immune-related genes that have been extensively studied in carcinogenicity were identified for stratification of immune microenvironment based on function and activity. The associations of highly TAGLN-related TFs with immune genes were analyzed in each classification using expression profile data and statistically analyzed for significance with the Limma package (version 3.34.8). Statistically significant genes in each group were presented in a scatter plot, and all correlations under each classification were generated using a heatmap using “plot” in the R language.
Correlation analysis between TAGLN and renowned genes related to cell viability and apoptosis
A collection of notable cancer genes that have been extensively studied in carcinogenicity were collected and classified into cell cycle-related and apoptosis-related mechanisms according to their function and activity. The apoptosis-related star genes were further categorized into G0-G1 phase and G2-M phase. The correlations between TAGLN and the star genes were estimated using the expression profile data for each class. Statistically significant genes in each group were analyzed using a scatter plot, and all correlations under each of the classifications were generated in a heatmap using “plot” in the R language.
Validation of the expression and distribution of skeletal protein transgelin in conditionally immortalized mouse podocytes
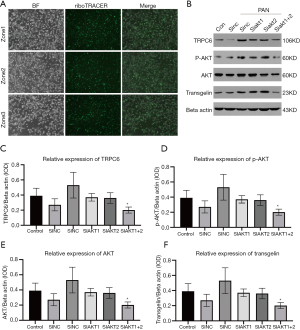
MPC-5 cells were inoculated in a 6-well plate at 2.5×105 cells per well with a total volume of 2 mL per well, and cultured in a 37 °C incubator for 24 h. The cells in the logarithmic growth phase were transfected with 15 siRNAs that were designed to specifically silence the expression of four genes AKT1, AKT2, PPAR alpha (phosphor S12), and TRPC6 (Figure S1), and mixed with Lipofectamine 3000 for sub-culturing 24 h, followed by PAN treatment for 24 h. The cells were trypsinized and then harvested and stained with DAPI. Representative images were obtained with a fluorescence microscope.
Harvested cells were lysed with 100 μL pre-cooled RIPA cell lysis buffer, homogenized with an electric homogenizer at 30,000 rpm for 10 min, and centrifuged at 13,000 rpm (4 °C) for 20 min. The supernatant was used for protein quantification with bicinchoninic acid (BCA) protein assay. Twenty microliter of protein lysate per well was loaded on gels for electrophoresis about 20 min, and proteins were transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was completely immersed in 5% BSA-TBST and incubated 1 h on a horizontal shaker. Primary antibodies AKT, p-AKT, transgelin, TRPC6 and Beta actin were diluted in 5% BSA-TBST and incubated overnight at 4 °C on a horizontal shaker. Secondary antibodies goat anti-rabbit IgG (H+L) HRP 1:10,000, rabbit anti-goat IgG (H+L) HRP 1:10,000, and goat anti-mouse IgG (H+L) HRP 1:10,000 were diluted in 5% BSA-TBST and incubated at room temperature for 40 min. Each PVDF membrane was rinsed with TBST-containing primary and secondary antibodies three times for 10 min, respectively. Specific proteins were detected by drop enhanced chemiluminescence (ECL) on the protein surface of the membrane to react for 3 min and exposure between 10 sec to 5 min. Band density was measured and normalized to Beta actin.
Results
Identification of DEGs
Dataset GSE108109 contains molecular targets and total RNA expression profile in the glomerular transcriptome from 62 NS, 23 AAV, one prototypical nephritic syndrome (PNS) and 6 healthy samples (living donor = control). 62 NS samples were divided into 14 MCD, 21 MGN and 25 FSGS. Log gene expression profile of TAGLN in 5 groups (NS, ANCA, MCD, MGN, FSGS and control) (Figure S2) were analyzed to identify a total of 1,987 DEGs from 10 comparisons, using |logFC|>1 and P<0.05 as the cutoff criteria (Table 1). Among them, 1,372 DEGs were detected in comparison of FSGS vs. control, among which 651 DEGs were upregulated and 721 were downregulated. 1,193 DEGs were identified between control and MCD, of which 545 DEGs were upregulated and 648 were downregulated. A total of 1,372 genes overlapped among DEGs from 10 comparisons for nephropathy were collected for STC with STEM (Figure S3), of which 877 DEGs were upregulated and 610 were downregulated (Table 1). In addition, 423 DEGs between FSGS and control were detected in GSE125779, among which 135 DEGs were upregulated and 288 were downregulated (Table 1).
Table 1
| Dataset | Compare | AllDifGene | UpGeneNum | DownGeneNum |
|---|---|---|---|---|
| GSE108109 | FSGS |
17 | 6 | 11 |
| GSE108109 | FSGS |
1,372 | 651 | 721 |
| GSE108109 | FSGS |
15 | 6 | 9 |
| GSE108109 | FSGS |
13 | 8 | 5 |
| GSE108109 | ANCAw |
1,301 | 557 | 744 |
| GSE108109 | ANCA |
37 | 20 | 17 |
| GSE108109 | ANCA |
60 | 49 | 11 |
| GSE108109 | MGN |
33 | 31 | 2 |
| GSE108109 | MCD |
1,193 | 545 | 648 |
| GSE108109 | MGN |
1,408 | 668 | 740 |
| GSE108109 | TotalDif | 1,987 | ||
| GSE108109 | SeriesCluster | 887 | 610 | |
| GSE125779 | FSGS |
423 | 135 | 288 |
| GSE99340 | hypoxia |
272 | 140 | 132 |
| GSE124622 | Adriamycin_ |
2,640 | 1,715 | 925 |
| GSE124622 | PAN_ |
9,096 | 5,686 | 3,410 |
A total of 272 DEGs between the normal oxygen group and hypoxia case group in GSE99340 were identified, using |logFC|>1 and P<0.05 as the cutoff criteria (Table 1). Among them, 140 DEGs such as heme oxygenase 1 (HMOX1), N-myc downstream regulated 1 (NDRG1) and podocyte cytoskeleton protein vinculin (VCL) were upregulated, and 132 DEGs including cytochrome P450 family 1 subfamily B member 1 (CYP1B1), vascular cell adhesion molecule 1 (VCAM1), interleukin 33 (IL33) and podocyte cytoskeleton proteins TAGLN and survivin (also called BIRC5) were downregulated. In addition, 1,246 DEGs between Adriamycin and control and 9096 between PAN and control were identified in GSE124622, respectively (Table 1). Cytoskeletal proteins ARHGAP24, TAGLN, TNNC1 and BIRC5 were downregulated, whereas RAC2, CPSF4, KRT7 and ARC were upregulated in comparison between Adriamycin and control. TAGLN and RAC2 were downregulated in comparison between MCD and control, but upregulated in comparison between FSGS and control. Genes KANK2, ACTN4, MYH9, INF2, ACTR2 and CDC42 were downregulated, whereas KANK4 and BIRC5 were upregulated in in comparison between PAN and control.
TAGLN tendency-dependent gene clustering in nephropathy
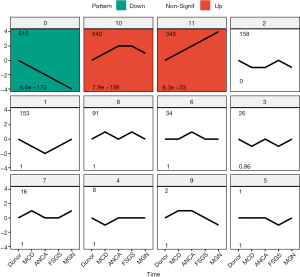
As shown Figure 1, according to STEM analysis, the DEGs were clustered into 12 important gene groups, which were ranked according to their significance (P value). Among 3 statistically significant clusters, the differences in the expression values of nearly all DEGs decreased in cluster 0 but increased in cluster 10 and 11 with the order of living donor (control), MCD, ANCA, FSGS, and MGN. Taken together, 610, 542 and 345 genes with significantly similar expression profiles to skeletal protein TAGLN were respectively assigned to group 0, 10 and 11 for subsequent candidate gene analysis.
GO and KEGG pathway enrichment analysis
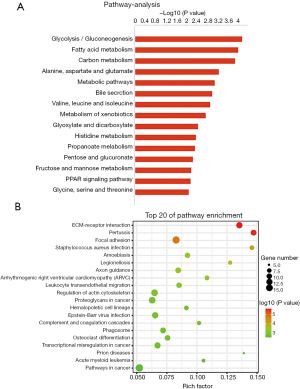
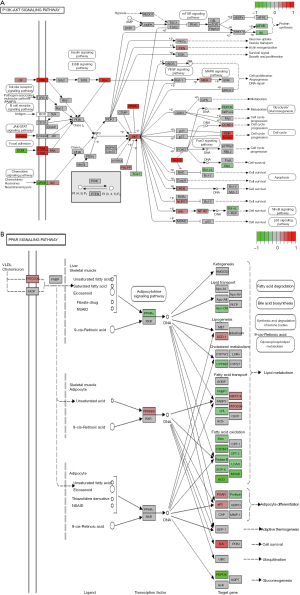
Overall, 850 overlapped genes among DEGs from GSE99340 and GSE108109 were collected and clustered into two groups based on Pearson correlation coefficients among the overlapped DEGs and TAGLN for KEGG analysis: significant positively TAGLN-related gene group or negatively TAGLN-related group. 369 DEGs that were significantly positively associated with TAGLN were enriched in 198 signal pathways, including P13k-AKT singling pathway, regulation of actin cytoskeleton, PARA singling pathway, ECM-receptor interaction, MPAK singling pathway, p53 singling pathway, NOD-like receptor signaling pathway, toll-like receptor (TLR) signaling pathway, tuberculosis, cytokine-cytokine receptor interaction and pathways in cancer. The top 20 pathways were shown in Figure 2A. A total of 169 DEGs were significantly negatively associated with TAGLN were enriched in 134 signal pathways, including P13k-Akt signaling pathway, regulation of actin cytoskeleton, PPAR signaling pathway, carbon metabolism, drug metabolism- cytochrome P450, glycolysis/gluconeogenesis and metabolic pathways. The top 20 pathways are shown in Figure 2B.
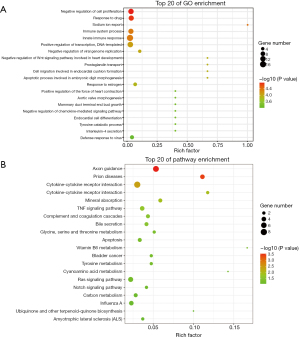
Seventy-six overlapped DEGs (25 upregulated and 41 downregulated) between GSE108109 and GSE125779 were associated with the innate immune response, positive regulation of transcription-DNA−templated, immune system process, negative regulation of cell proliferation, response to drug, and defense response to virus (Figure 3A). The overlapped genes were enriched in the important signal pathways, including cytokine−cytokine receptor interaction, Ras signaling pathway, axon guidance, TNF signaling pathway and apoptosis (Figure 3B).
Co-expression interactive network analysis of transgelin-related DEGs
The interactive network among the 495 DEGs were determined using STRING to reveal the coexpression, genetic, and physical interactions among the DEGs and predicted genes. The network of 27 positive TAGLN -related genes is shown in Figure 4A. The 20 top hub nodes with the highest degrees of connectivity included transcription factor p65 (RELA), Tumor Protein P53 (TP53), Rac family small GTPase 2 (RAC2), Alpha-actinin 1(ACTN1), placental growth factor (PGF), EPH Receptor A2 (EPHA2), SHC Adaptor Protein 1 (SHC1), colony stimulating factor 1 receptor (CSF1R), SHC Adaptor Protein 4 (SHC4), platelet derived growth factor Subunit B (POGFB), Collagen Type I Alpha 2 Chain (COL1A2), Collagen Type III Alpha 1 Chain (COL3A1), Integrin Subunit Alpha (ITGA) 3/4/5/11, Collagen Type IV Alpha 2 Chain (COL4A2), Cluster of Differentiation (CD) 44, fibronectin 1(FN1), and Transcription factor PU.1 (SPI1).
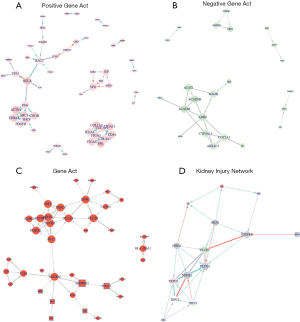
The network of 54 negative TAGLN-related genes is shown in Figure 4B. The ten top hub nodes with the highest degrees of connectivity were acetoacetyl-CoA thiolase (ACAT1), short/branched chain specific acyl-CoA dehydrogenase (ACADSB), Acyl-CoA Dehydrogenase Medium Chain (ACADM), Alcohol dehydrogenase 6 (ADH6), Monoamine oxidase B (MAOB), Cytochrome P450 4A11 (CYP4A11), UDP-glucuronosyltransferase 2A3 (UGT2A3), Aldo-keto reductase family 1 member C3 (AKR1C3), Fructose-bisphosphate aldolase B (ALDOB) and Fructose-1,6-bisphosphatase 1 (FBP1). TAGLN-regulating histone acetylation appears to play an important role in structural and functional changes of podocytes and proteinuria.
GSE35488 is from the platform GPL14663 with human expression data with IgA nephropathy (IgAN) [HG-U133A_ENTREZG_10] from 21 patients with IgAN. The PPI relationships showed that COL1A1, EGR1, ELF3, IER3, HBEGF, MAFF, MCL1, SAMD4A, SERPINE1, STEAP1 and TYMS were the hub genes with the highest degrees of connectivity (Figure 4C).
The co-expressions among TAGLN and TRPC family proteins F2R, PLCE1 and TRPC 3/5/6 were estimated with Pearson correlation coefficient, and interactive expression network was developed using the “ggplot2” package. Expression of transgelin was significantly positively associated with F2R. As shown in Figure 4D, TAGLN interacted with the TRPC signaling pathway by regulating platelet derived growth factor receptor beta (PDGFRB) to control many important processes in the cell such as alteration of intracellular calcium concentration.
The correlation analysis between TAGLN and podocyte injury-related genes
A collection of notable genes associated with cell cycle were classified as G0-G1 phase and G2-M phase according to their function. The correlations of TAGLN expression profile with expression profiling of cell cycle-related genes Catenin Delta 1 (CDND1), Cyclin Dependent Kinase (CDK) 2/6, Cyclin Dependent Kinase Inhibitor 1A (CDKN1A) and TP53 were estimated in G0-G1 and G2-M stage, respectively. TAGLN expression was significantly positively related to CDND1, CDK2 and CDK6 (P<0.05) in G0-G1 phase, and significantly positively associated with CDKN1A and TP53, but negatively with CDKN1B (P<0.05) in G2-M phase (Figure S4A). TAGLN expression was significantly positively associated with apoptosis-related renowned genes TP53, poly ADP-ribose polymerase 1 (PARP1), but negatively with B-cell lymphoma-2 (BCL2; P<0.05) (Figure S4B).
The correlation analysis between TAGLN and immunity
Sixteen genes overlapped between TFs and DEGs in GSE99340 and GSE108109 (Figure S5), from which 12 TFs were significantly associated with Transgelin (Table 2). The associations of 11 TFs-regulating transgelin with the proven immune cells and immune factors, cytokines and chemokines are shown in Figure 5. TAGLN was significantly positively associated with Integrin alpha M (ITGAM), CD68, CSF1R and CD163 in the immune microenvironment of Tyros, Axl and Mer (TAM) (Figure 5A), and with ITGAM in neutrophils (Figure 5B). TAGLN-related TF SPI1 was significantly associated with a majority of immune cells and immune factors, cytokines and chemokines in each immune microenvironment (Figure 5A,D,F,H). CD4 was significantly related with TAGLN and all TAGLN-related TFs in lymphoid lineage (Figure 5E), Helper (Th) cell (Figure 5D) and Treg cell (Figure 5M). CD14 was significantly associated with the most TAGLN and all TAGLN-related TFs in TEM (Figure 5L), MDSC (Figure 5F) and myeloid lineage (Figure 5D); whereas CSF1R was significantly associated with the most of TAGLN and all TAGLN-related TFs in TAM (Figure 5A) and myeloid lineage (Figure 5D) immune microenvironment.
Table 2
| TF | Correlation with TAGLN | P_value | P_adj | Function |
|---|---|---|---|---|
| DRAP1 | 0.2560 | 0.0067 | 0.0145 | DR1-associated protein 1 (negative cofactor 2 alpha) |
| HLF | −0.4499 | 0.0000 | 0.0000 | Hypermethylated in cancer 1 |
| HNF4G | −0.4239 | 0.0000 | 0.0001 | Hepatocyte nuclear factor 4, gamma |
| MNT | 0.2675 | 0.0045 | 0.0107 | MAX network transcriptional repressor |
| NFATC1 | 0.2691 | 0.0043 | 0.0103 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| POLR2A | 0.2274 | 0.0164 | 0.0295 | Polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| PPARGC1A | −0.3511 | 0.0002 | 0.0008 | Peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| RARA | 0.2635 | 0.0052 | 0.0119 | Retinoic acid receptor, alpha |
| SKI | 0.2895 | 0.0021 | 0.0060 | SKI proto-oncogene |
| SPI1 | 0.2926 | 0.0018 | 0.0055 | Spi-1 proto-oncogene |
| TEAD3 | 0.3807 | 0.0000 | 0.0003 | TEA domain family member 1 (SV40 transcriptional enhancer factor) |
Western blotting analysis of selected genes
To directly test the role of DEGs in TAGLN expression, MPC-5 cells were transfected with 15 siRNAs that were designed to specifically silence the expression of four DEGs to knock down TAGLN: AKT1, AKT2, PPAR alpha and TRPC6. As shown in Figure 6A, MPC-5 cells were successfully transfected with plasmids with transfection efficiency of 70–80%. For all 4 DEGs, no difference in protein expression was observed between the siRNA NC group (empty plasmid) and the control group (Figure 6B-F). In the siRNA NC group, transfection with siRNA NC and PAN treatment increased protein expression of TRPC6, and phosphorylation level of p-AKT. In the siAKT1 group, no significant change in protein expression of TRPC6 and phosphorylation level of p-AKT was observed by transfecting with AKT1 siRNA and siAKT2, and treating with PAN. Transfection of AKT1 siRNA and AKT2 siRNA and PAN treatment significantly reduced relative expression of TRPC6, p-AKT, AKT and transgelin, suggesting that simultaneous transfection of AKT1 siRNA and AKT2 siRNA inhibited effects of PAN on protein expression of TRPC6, p-AKT, AKT and transgelin.
Discussion
The actin cytoskeleton plays an important role in immune function in coordination with numerous cellular processes including cell proliferation and differentiation, apoptosis, migration, and cellular signaling (29,31) by spatially and temporally regulating actin filaments with actin-binding proteins (ABPs). Transgelin is encoded by the gene TAGLN and belongs to the transgelin superfamily of actin cross-linking/gelling proteins, acting as a transformation-sensitive and shape change-sensitive ABP in actin dynamics and actin-associated signaling pathways to affect cell motility (17,18,22) and immunity (32,33). In the present study, GO and KEGG analysis revealed that DEGs were primarily associated with innate immunity, positive regulation of transcription-DNA−templated, immune system processes and negative regulation of cell proliferation. DEGs were also involved in the enrichment of cytokine-cytokine receptor interaction signaling pathway, Ras signaling pathway, Axon guidance, TNF signaling pathway and apoptosis. TAGLN and TAGLN-related TFs were demonstrated to participate in interactions with the notable genes related to key cellular processes such as cell proliferation and differentiation and apoptosis, and modulation of immune cells and immune factors, cytokines, and chemokines in different immune microenvironments. TAGLN expression was significantly positively associated with TP53 and PARP1, but negatively with BCL2, supporting the results of Zhang et al. (2010) (34), who reported activation of the mitochondria-associated apoptosis pathway by transfecting with transgelin in LNCaP cells. Transgelin-induced p53 expression affects the subcellular distribution and cytoplasmic accumulation of TP53 (35), resulting in the downregulation of BCL2 to activate caspase-3. Zhou et al. (2016) reported that transgelin overexpression in isogenic colorectal cancer cell lines altered the levels of approximately 250 other transcripts involved in cell motility such as increased expression of HOOK1, SDCCAG8, ENAH/Mena, and TNS1, and decreased representation of EMB, BCL11B, and PTPRD in vivo, leading to function alternation of actin or other cytoskeletal proteins (35). The reactive oxygen species (ROS)-mediated NF-κB pathway is activated by transgelin downregulation, inducing proinflammatory vascular smooth muscle cells (32,36). As the key player in the PI3K/Akt signaling pathway (37), the NF-κB pathway might be the upstream regulating pathway for transgelin. Sod2 expression was enhanced by TAGLN knockdown to activate p47 phox, reflecting contributions of mitochondria and NADPH oxidase to augment ROS production (32), which might be caused by actin and microtubule cytoskeletal remodeling. Transgelin expression has been widely studied at the transcriptional, translational, and post-translational levels in response to various intercellular events; however, regulatory mechanisms of transgelin in immunity and cellular processes are largely unknown. Further study is needed to validate interactions of transgelin or TAGLN-related TFs with immunity and cellular processes in the progression of proteinuria and podocyte injury.
Our study demonstrated positive associations of increased TRPC expression in podocytes with nephropathy and proteinuria, suggesting an important role of TRPC in TAGLN- regulating actin cytoskeleton in podocytes (38,39). TRPC, including TRPC1, TRPC3, TRPC4, TRPC5 and TRPC6, is a member of a family of proteins that express in podocytes (40), regulate Ca2+ influx into podocytes (38), and play the key role in the pathogenesis of podocyte injury and proteinuria (39). TRPC6 interacts with nephrin and podocin in the slit membrane by localizing in the podocyte cell body and primary processes. TRPC6 is downregulated by high glucose, leading to impaired Ca2+ signaling of mesangial cells (41). Gain-of-function TRPC6 mutations were discovered in families with an autosomal dominant form of FSGS ions, leading to familial FSGS (42). This condition was enhanced by the influx of Ca2+, particularly after the activation of the G-protein-coupled receptor (GPCR) AT1 by angiotensin II (Ang II). In addition, the increased levels of wild-type TRPC6 protein in some acquired glomerular diseases, such as membranous nephropathy and PAN-induced albuminuria, might result in podocyte dysfunction (43). Ang II has been shown to contribute to podocyte injury by increasing TRPC6 expression via a nuclear factor of activated T-cells (NFAT)-mediated positive feedback signaling pathway (39). TRPC5 acts as a mediator of cytoskeletal changes in podocytes by inducing Rac1 activation and synaptopodin degradation (44,45). Opposite effects of TRPC5 and TRPC6 on podocyte actin dynamics and cell motility (43,44) have been demonstrated by regulating RhoA and Rac1, respectively. In the present study, TRPC6 knockdown led to significantly reduced protein expression of TAGLN, suggesting a significant role in the pathogenesis of podocyte injury and proteinuria by regulating RhoA and Rac1 in PI3K-AKT signaling pathway and PPAR signaling pathway to control many key cellular processes, including alternation of intracellular calcium concentration.
Conclusions
Our study demonstrated that podocytes responded to injury and proteinuria along defined pathways. The top DEGs-enriched pathways included PI3K-AKT signaling pathway, PPAR signaling pathway, and glycolysis/gluconeogenesis and metabolic pathways. TAGLN significantly positively interacted with 78 genes including PDGFA, FN1, RELA, THBS3, COL1A2, VEGFC, OSMR, LAMC1 and EPHA2, and negatively interacted with 19 genes such as PRLR, PCK1/2, EGF, FGFR2, GHR and INSR in PI3K-AKT signaling pathway (Figure 7A). In the present study, TAGLN knockdown led to significantly reduced protein expression of TRPC6 and TAGLN, as well as phosphorylation level of p-AKT in the PI3K-AKT signaling pathway. As a second messenger related to intracellular signal transduction, PI3K activates Akt to play a major role in tolerance to podocyte apoptosis by the PI3K-Akt signaling pathway (Figure 7A). Proteinuria is caused by podocyte injury induced by purine adenosine, AngII, TGF-β1, protein overload and hemodynamic disorders (46). The insulin-like growth factor 1 receptor β (IGF1Rβ)/PI3K/AKT pathway is activated by TAGLN2, a homolog of TAGLN through recruiting focal adhesion kinase to the IGF1R signaling complex (47). Podocyte injury is promoted by microRNA-27a to activate PPARγ-mediated β-catenin in diabetic nephropathy (48) but prevented by PPAR-γ–mediated TRPC6 inhibition (49). In the present study, TAGLN interacted with the PPAR signaling pathway, including nine upregulated genes such as ILK, CD36, SLC27A5, ANGPTL4 and FABP4, and 17 downregulated genes such as ACADM, ACOX2, PCK1/2, SLC27A2, CYP8B1 and ACAA1 (Figure 7B). In addition, glycolysi/gluconeogenesis and metabolic pathways were involved in occurrence and development of podocyte injury and proteinuria. Podocytes are also covered by glycocalyx, principally podocalyxin, a sialomucin closely related to CD34 and endoglycan. Podocalyxin was demonstrated to regulate adhesion and cell morphology by interactions with several intracellular proteins and at least one extracellular ligand, playing an important role in the formation and maintenance of podocyte foot processes in the developing kidney (50). In addition, many hub genes were characterized by the PPI network construction for the DEGs with the top 10 as COL1A1, EGR1, ELF3, IER3, HBEGF, MAFF, MCL1, SAMD4A, SERPINE1, STEAP1 and TYMS. The extrapolation of these findings to humans is limited without validation in real-world clinical samples of Chinese patients. Future studies are needed to validate the results of the present study and understand the roles of these important hub genes in progression of podocyte injury and proteinuria.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 81400718), Research Foundation of Beijing Friendship Hospital, Capital Medical University (Grant No. yyqdkt2018-21), and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding (Grant No. ZYLX201824).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-83
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-83). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bello AK, Hemmelgarn B, Lloyd A, et al. Associations among estimated glomerular filtration rate, proteinuria, and adverse cardiovascular outcomes. Clin J Am Soc Nephrol 2011;6:1418-26. [Crossref] [PubMed]
- Merscher S. Editorial: Molecular Mechanisms of Proteinuria. Front Med (Lausanne) 2018;5:300. [Crossref] [PubMed]
- Perico L, Conti S, Benigni A, et al. Podocyte-actin dynamics in health and disease. Nat Rev Nephrol 2016;12:692-710. [Crossref] [PubMed]
- Müller-Deile J, Schiffer M. Podocytes from the diagnostic and therapeutic point of view. Pflugers Arch 2017;469:1007-15. [Crossref] [PubMed]
- Drenckhahn D, Franke RP. Ultrastructural organization of contractile and cytoskeletal proteins in glomerular podocytes of chicken, rat, and man. Lab Invest 1988;59:673-82. [PubMed]
- Andrews PM, Bates SB. Filamentous actin bundles in the kidney. Anat Rec 1984;210:1-9. [Crossref] [PubMed]
- Kobayashi N. Mechanism of the process formation; podocytes vs. neurons. Microsc Res Tech 2002;57:217-23. [Crossref] [PubMed]
- Lennon R, Randles MJ, Humphries MJ. The importance of podocyte adhesion for a healthy glomerulus. Front Endocrinol (Lausanne) 2014;5:160. [Crossref] [PubMed]
- Schell C, Huber TB. The Evolving Complexity of the Podocyte Cytoskeleton. J Am Soc Nephrol 2017;28:3166-74. [Crossref] [PubMed]
- Cil O, Perwad F. Monogenic Causes of Proteinuria in Children. Front Med (Lausanne) 2018;5:55. [Crossref] [PubMed]
- Kaplan JM, Kim SH, North KN, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 2000;24:251-6. [Crossref] [PubMed]
- Brown EJ, Schlöndorff JS, Becker DJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 2010;42:72-6. [Crossref] [PubMed]
- Mele C, Iatropoulos P, Donadelli R, et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med 2011;365:295-306. [Crossref] [PubMed]
- Kim JM, Wu H, Green G, et al. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science 2003;300:1298-300. [Crossref] [PubMed]
- Miao J, Fan Q, Cui Q, et al. Newly identified cytoskeletal components are associated with dynamic changes of podocyte foot processes. Nephrol Dial Transplant 2009;24:3297-305. [Crossref] [PubMed]
- Yu H, Königshoff M, Jayachandran A, et al. Transgelin is a direct target of TGF-beta/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J 2008;22:1778-89. [Crossref] [PubMed]
- Elsafadi M, Manikandan M, Dawud RA, et al. Transgelin is a TGFβ-inducible gene that regulates osteoblastic and adipogenic differentiation of human skeletal stem cells through actin cytoskeleston organization. Cell Death Dis 2016;7:e2321 [Crossref] [PubMed]
- Yamamura H, Masuda H, Ikeda W, et al. Structure and expression of the human SM22alpha gene, assignment of the gene to chromosome 11, and repression of the promoter activity by cytosine DNA methylation. J Biochem 1997;122:157-67. [Crossref] [PubMed]
- Shen X, Jiang H, Ying M, et al. Calcineurin inhibitors cyclosporin A and tacrolimus protect against podocyte injury induced by puromycin aminonucleoside in rodent models. Sci Rep 2016;6:32087. [Crossref] [PubMed]
- Dong N, Meng L, Xue R, et al. Adrenomedullin ameliorates podocyte injury induced by puromycin aminonucleoside in vitro and in vivo through modulation of Rho GTPases. Int Urol Nephrol 2017;49:1489-506. [Crossref] [PubMed]
- Zhang H, Luo W, Sun Y, et al. Wnt/β-Catenin Signaling Mediated-UCH-L1 Expression in Podocytes of Diabetic Nephropathy. Int J Mol Sci 2016;17:1404. [Crossref] [PubMed]
- Ogawa A, Sakatsume M, Wang X, et al. SM22alpha: the novel phenotype marker of injured glomerular epithelial cells in anti-glomerular basement membrane nephritis. Nephron Exp Nephrol 2007;106:e77-87. [Crossref] [PubMed]
- Hauser PV, Perco P, Mühlberger I, et al. Microarray and bioinformatics analysis of gene expression in experimental membranous nephropathy. Nephron Exp Nephrol 2009;112:e43-58. [Crossref] [PubMed]
- Gong W, Yu J, Wang Q, et al. Estrogen-related receptor (ERR) γ protects against puromycin aminonucleoside-induced podocyte apoptosis by targeting PI3K/Akt signaling. Int J Biochem Cell Biol 2016;78:75-86. [Crossref] [PubMed]
- Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 2006;7:191. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-8. [Crossref] [PubMed]
- Kumar MS, Adki KM. Marine natural products for multi-targeted cancer treatment: A future insight. Biomed Pharmacother 2018;105:233-45. [Crossref] [PubMed]
- Comrie WA, Burkhardt JK. Action and Traction: Cytoskeletal Control of Receptor Triggering at the Immunological Synapse. Front Immunol 2016;7:68. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Fritzsche M, Fernandes RA, Chang VT, et al. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci Adv 2017;3:e1603032 [Crossref] [PubMed]
- Shen J, Yang M, Ju D, et al. Disruption of SM22 promotes inflammation after artery injury via nuclear factor kappaB activation. Circ Res 2010;106:1351-62. [Crossref] [PubMed]
- Jo S, Kim HR, Mun Y, et al. Transgelin-2 in immunity: Its implication in cell therapy. J Leukoc Biol 2018;104:903-10. [Crossref] [PubMed]
- Zhang ZW, Yang ZM, Zheng YC, et al. Transgelin induces apoptosis of human prostate LNCaP cells through its interaction with p53. Asian J Androl 2010;12:186-95. [Crossref] [PubMed]
- Zhou HM, Fang YY, Weinberger PM, et al. Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility. BMC Cancer 2016;16:55. [Crossref] [PubMed]
- Lees-Miller JP, Heeley DH, Smillie LB. An abundant and novel protein of 22 kDa (SM22) is widely distributed in smooth muscles. Purification from bovine aorta. Biochem J 1987;244:705-9. [Crossref] [PubMed]
- Francés R, Tumang JR, Kaku H, et al. B-1 cells express transgelin 2: unexpected lymphocyte expression of a smooth muscle protein identified by proteomic analysis of peritoneal B-1 cells. Mol Immunol 2006;43:2124-9. [Crossref] [PubMed]
- Ilatovskaya DV, Staruschenko A. TRPC6 channel as an emerging determinant of the podocyte injury susceptibility in kidney diseases. Am J Physiol Renal Physiol 2015;309:F393-7. [Crossref] [PubMed]
- Nijenhuis T, Sloan AJ, Hoenderop JG, et al. Angiotensin II contributes to podocyte injury by increasing TRPC6 expression via an NFAT-mediated positive feedback signaling pathway. Am J Pathol 2011;179:1719-32. [Crossref] [PubMed]
- Kalwa H, Storch U, Demleitner J, et al. Phospholipase C epsilon (PLCε) induced TRPC6 activation: a common but redundant mechanism in primary podocytes. J Cell Physiol 2015;230:1389-99. [Crossref] [PubMed]
- Riccio A, Medhurst AD, Mattei C, et al. mRNA distribution analysis of human TRPC family in CNS and peripheral tissues. Brain Res Mol Brain Res 2002;109:95-104. [Crossref] [PubMed]
- Hofstra JM, Lainez S, van Kuijk WH, et al. New TRPC6 gain-of-function mutation in a non-consanguineous Dutch family with late-onset focal segmental glomerulosclerosis. Nephrol Dial Transplant 2013;28:1830-8. [Crossref] [PubMed]
- Möller CC, Wei C, Altintas MM, et al. Induction of TRPC6 channel in acquired forms of proteinuric kidney disease. J Am Soc Nephrol 2007;18:29-36. [Crossref] [PubMed]
- Schaldecker T, Kim S, Tarabanis C, et al. Inhibition of the TRPC5 ion channel protects the kidney filter. J Clin Invest 2013;123:5298-309. [Crossref] [PubMed]
- Tian D, Jacobo SM, Billing D, et al. Antagonistic regulation of actin dynamics and cell motility by TRPC5 and TRPC6 channels. Sci Signal 2010;3:ra77. [Crossref] [PubMed]
- Saleem MA. One hundred ways to kill a podocyte. Nephrol Dial Transplant 2015;30:1266-71. [Crossref] [PubMed]
- Kim IG, Lee JH, Kim SY, et al. Hypoxia-inducible transgelin 2 selects epithelial-to-mesenchymal transition and γ-radiation-resistant subtypes by focal adhesion kinase-associated insulin-like growth factor 1 receptor activation in non-small-cell lung cancer cells. Cancer Sci 2018;109:3519-31. [Crossref] [PubMed]
- Zhou Z, Wan J, Hou X, et al. MicroRNA-27a promotes podocyte injury via PPARγ-mediated β-catenin activation in diabetic nephropathy. Cell Death Dis 2017;8:e2658 [Crossref] [PubMed]
- Sonneveld R, Hoenderop JG, Isidori AM, et al. Sildenafil Prevents Podocyte Injury via PPAR-γ-Mediated TRPC6 Inhibition. J Am Soc Nephrol 2017;28:1491-505. [Crossref] [PubMed]
- Nielsen JS, McNagny KM. The role of podocalyxin in health and disease. J Am Soc Nephrol 2009;20:1669-76. [Crossref] [PubMed]


