
The potential mechanism of Astragali Radix in the treatment of children with nephrotic syndrome
Introduction
Nephrotic syndrome (NS), whose clinical features mainly include proteinuria, hypoproteinemia, hyperlipidemia, and systemic edema, is one of the most common renal diseases in children (1). The pathogenesis of NS is very complex, and is mainly related to podocyte injury, apoptosis, lipid metabolism disorder, and oxidative stress (2). At present, the treatment of NS is still dominated by chemical drugs; however, these drugs can cause serious toxic and side effects and complications (3). Conversely, traditional Chinese medicine considers overall regulation of the body and has less toxic and side effects than western medicine.
Studies have shown that Fangji Huangqi Decoction, Angelica Tonifying Blood Decoction, and Shenqi Dihuang Decoction all use Astragali Radix as the main drug, which has significant efficacy in the treatment of nephropathy (4,5). Astragali Radix originated from “Shennong’s Herbal Classic” in the eastern Han Dynasty. As a perennial legume herb, it is based on Astragalus Mongol and Astragalus membranaceus. Astragali Radix has a sweet taste and warm nature, and is an important traditional Chinese medicine with a history of more than 2,000 years (6,7). Recent studies have found that astragalus can reduce urinary protein excretion and reduce kidney injury (8,9). Clinical studies have found (10) that the oxidative stress of NS patients is enhanced, and astragalus membranaceus has certain antioxidant effects. The efficacy of astragalus in NS intervention is relatively clear; however, its molecular mechanism is still unclear, and thus further research and exploration is needed. This study took common human disease targets as the research objects and explored the mechanism of Astragali Radix in the treatment of NS based on reverse molecular docking technology.
Due to the multi-component, multi-pathway, and multi-target characteristics of traditional Chinese medicine, it is difficult to clarify the substance basis and mechanism of action of traditional Chinese medicine. Many scholars have tried to explain the mechanism of action of traditional Chinese medicine with modern medicine. The concept of “network pharmacology” was first proposed by the British pharmacologist Hopkins in 2007. Based on the multidisciplinary theories, such as systems biology and multidimensional pharmacology, the molecular mechanism of drug intervention in diseases can be understood from a multidimensional perspective (11). Previous studies have shown that the study of mechanism of action of Chinese medicine through network pharmacology is in line with the overall action characteristics of Chinese medicine, and the method is accurate and reliable. For example, some scholars have used network pharmacological methods to study the mechanism of Naozhenning Granules’ intervention for brain injury (12), the anti-depressive mechanism of Bubuurum (13), and the mechanism of Scolt Flower’s in relieving cough and reducing phlegm (14). Based on the 20 components of Astragali Radix, this study used the network pharmacology method to construct a component-target-disease network, and explored the mechanism of action of the multiple components, multiple targets, and multiple pathways of Astragali Radix in the treatment of NS to pave the way for further research on the mechanism of action of Astragali Radix in the treatment of NS. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-348).
Methods
Collection and screening of chemical constituents of Astragali Radix
The chemical composition of Astragali Radix was obtained from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (http://lsp.nwu.edu.cn/tcmsp.php) (15). We input the keyword “Astragali Radix” to obtain information on its active ingredients. In this study, oral bioavailability (OB) and drug-likeness (DL) were used to screen out the chemical components with an OB ≥30% and a DL ≥0.18.
Forecast potential targets
The PharmMapper Database (http://59.78.98.102/pharmmapper/index.php) was used to obtain the mol2 format file of the candidate components of Astragali Radix, and the calculation results were then downloaded. The Protein Data Bank identification (PDB ID), target name, and fit score were obtained after submission. The fit score refers to the matching degree between the molecule and target; the higher the score, the better the match. Target proteins with a fit score >3 were selected as the target proteins of the compound, and the PDB ID of the protein target was input into the Uniprot Database (http://www.uniprot.org/) to obtain the potential action targets of the main components of Astragali Radix using retrieval and transformation operation.
The keyword “Nephrotic syndrome” was input in the Online Mendelian Inheritance in Man (OMIM; https://omim.org) and GeneCards (https://www.genecards.org/) databases to search for genes related to NS. Duplicate genes and false positive genes were removed, and the remaining genes were matched with the above targets to screen those related to the active components of Astragali Radix.
Construction and analysis of the protein-protein-interaction network
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (16) (https://string-db.org/) collects details of a large number of protein-interaction relationships. By importing the protein targets of Astragali Radix into the STRING Database and defining the species as human, a protein-protein-interaction analysis was carried out. Node1, node2, and the combined score information were imported from the exported file into Cytoscape Software. The node size was set to reflect the degree value, and the thickness of the edge was set to reflect the size of the combined score to produce the final protein-interaction network.
Analysis of biological processes and pathways
The action targets of Astragali Radix were then imported into the DAVID database, the select identifier was set as the “official gene symbol,” the list type was set as the “gene list,” and the species were defined as human. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed on Astragali Radix action targets, and the results were saved. A threshold of P<0.05 was set, and the number of targets involved was sorted according to the number of targets. The biological processes or pathways with the top rankings were screened and mapped with EXCEL. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
Most of the analyses were done automatically in the database. Graphics are made by Excel or automatically generated by database. All statistical analyses were two-sided, with a P value <0.05 considered statistically significant.
Results
Active components of Astragali Radix
In the TCMSP database, 20 chemical constituents were screened according to the OB and DL values. The basic information of the 20 components are summarized in Table 1. The target network diagram of the Astragali Radix single drug is shown in Figure 1.
Table 1
| Serial number | MOLID | Relative molecular weight | Compound | OB/% | DL | AlgP |
|---|---|---|---|---|---|---|
| 1 | MOL000211 | 456.78 | Mairin | 55.37707338 | 0.7761 | 6.521 |
| 2 | MOL000239 | 314.31 | Jaranol | 50.82881677 | 0.29148 | 2.087 |
| 3 | MOL000296 | 414.79 | Hederagenin | 36.91390583 | 0.75072 | 8.084 |
| 4 | MOL000033 | 428.82 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | 36.22847056 | 0.78288 | 8.54 |
| 5 | MOL000354 | 316.28 | Isorhamnetin | 49.60437705 | 0.306 | 1.755 |
| 6 | MOL000371 | 314.36 | 3,9-di-O-methylnissolin | 53.74152673 | 0.47573 | 2.892 |
| 7 | MOL000374 | 642.67 | 5’-hydroxyiso-muronulatol-2’,5’-di-O-glucoside | 41.71766574 | 0.69251 | −0.948 |
| 8 | MOL000378 | 316.38 | 7-O-methylisomucronulatol | 74.68613752 | 0.29792 | 3.379 |
| 9 | MOL000379 | 462.49 | 9,10-dimethoxypterocarpan-3-O-13-dimethyl-1 | 36.73668801 | 0.9243 | 0.737 |
| 10 | MOL000380 | 300.33 | (6ar,11ar)-9,10-dimethoxy-6a,11a-dihydro-6H-benzofurano[3,2-c]chromen-3-ol | 64.25545452 | 0.42486 | 2.641 |
| 11 | MOL000387 | 418.38 | Bifendate | 31.09782391 | 0.66553 | 2.563 |
| 12 | MOL000392 | 268.28 | Formononetin | 69.67388061 | 0.21202 | 2.583 |
| 13 | MOL000398 | 316.33 | Isoflavanone | 109.9866565 | 0.29572 | 2.415 |
| 14 | MOL000417 | 284.28 | Calycosin | 47.75182783 | 0.24278 | 2.316 |
| 15 | MOL000422 | 286.25 | Kaempferol | 41.88224954 | 0.24066 | 1.771 |
| 16 | MOL000433 | 441.45 | Fa | 68.96043622 | 0.7057 | 0.007 |
| 17 | MOL000438 | 302.35 | (3R)-3-(2-hydroxy-3,4-dimethoxyphenyl)chroman-7-ol | 67.66747949 | 0.26479 | 3.128 |
| 18 | MOL000439 | 626.67 | Isomucronulatol-7,2’-di-O-glucosiole | 49.28105539 | 0.62065 | −0.68 |
| 19 | MOL000442 | 314.31 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene | 39.04541112 | 0.47943 | 3.113 |
| 20 | MOL000098 | 302.25 | Quercetin | 46.43334812 | 0.27525 | 1.504 |
Target prediction
All the targets obtained from the PharmMapper database were collected. After removing the duplicates and false positive genes, a total of 394 targets of active ingredients of Astragali Radix remained. By comparing the NS-related genes in the GeneCards and OMIM databases, a total of 39 potential NS-related targets were finally identified (see Table 2).
Table 2
| Serial number | Uniprot | Targets |
|---|---|---|
| 1 | Q06124 | PTPN11 |
| 2 | P15056 | BRAF |
| 3 | P02768 | ALB |
| 4 | P01112 | HRAS |
| 5 | Q92793 | CREBBP |
| 6 | P42768 | WAS |
| 7 | P31749 | AKT1 |
| 8 | P06213 | INSR |
| 9 | P12821 | ACE |
| 10 | P61769 | B2M |
| 11 | P02766 | TTR |
| 12 | P00492 | HPRT1 |
| 13 | P01009 | SERPINA1 |
| 14 | P10721 | KIT |
| 15 | P04150 | NR3C1 |
| 16 | P98170 | XIAP |
| 17 | O60674 | JAK2 |
| 18 | P00533 | EGFR |
| 19 | P37231 | PPARG |
| 20 | P00734 | F2 |
| 21 | P02753 | RBP4 |
| 22 | P43403 | ZAP70 |
| 23 | P00797 | REN |
| 24 | P07359 | GP1BA |
| 25 | P60568 | IL2 |
| 26 | P05019 | IGF1 |
| 27 | P08253 | MMP2 |
| 28 | P28482 | MAPK1 |
| 29 | P35221 | CTNNA1 |
| 30 | Q00987 | MDM2 |
| 31 | P60953 | CDC42 |
| 32 | P13726 | F3 |
| 33 | P29474 | NOS3 |
| 34 | P09871 | C1S |
| 35 | P61626 | LYZ |
| 36 | P14780 | MMP9 |
| 37 | P08473 | MME |
| 38 | P42224 | STAT1 |
| 39 | P04035 | HMGCR |
Protein-protein-interaction network
The selected targets were input into the String Database, the species were limited to human, and the results were imported into Cytoscape Software to obtain the interaction network among the targets (see Figure 2). The nodes represent proteins, and the edges represent associations between proteins. The network comprised a total of 39 nodes and 366 edges. The size of a node in Figure 2 represents the size of the degree value; the larger the size of a node, the larger the degree. The thickness of the edge represents the combined score; the thicker the edge, the greater the combined score. The top 5 proteins were albumin (ALB), serine/threonine kinase (AKT1), epidermal growth factor receptor (EGFR), mitogen-activated protein kinase (MAPK), and matrix metallopeptidase 9 (MMP 9).

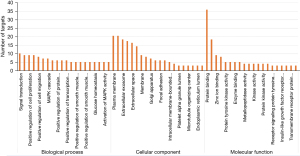
Gene function and pathway analysis
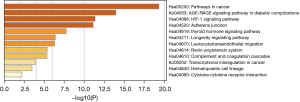
A GO analysis and a KEGG analysis were performed on the 39 action targets predicted by Astragali Radix active components by the DAVID database. As Figure 3 shows, the GO rich-set analysis included biological processes, cellular components, and molecular functions. The biological processes included signal transduction, the positive regulation of cell proliferation, the positive regulation of migration, and the MAPK cascade. Cell composition included the plasma membrane, extracellular exosome, and extracellular space. The molecular functions included protein binding, zinc-ion binding, protein tyrosine kinase activity, and enzyme binding. The results of the pathway analysis are shown in Figure 4. The treatment of NS by Astragali Radix mainly involved pathways in cancer, proteoglycans in cancer, the phosphatidylinositol 3-kinase and protein kinase B (PI3K-Akt) signaling pathway, the rennin-angiotensin-system (Ras) signaling pathways, and Forkhead box protein O1 (FoxO) signaling pathways.
Discussion
Astragali Radix is an important drug in the treatment of NS, which has the effects of diuresis and detumescence. The compounds in which Astragali Radix is the main drug account for a vast proportion of NS treatments. Thus, this study on the treatment of NS with Astragali Radix is of great significance. There was a similar report about the mechanism of Astragali Radix in the treatment of children with nephrotic syndrome (17), however, our study used more data from official platforms, such as TCMSP, OMIM, and DAVID database, and so the results were relatively more reliable.
A reverse molecular docking method was used to predict the action targets of active ingredients, and combined with the relevant database, 39 NS-related action targets were identified. Among them, matrix metalloproteinases (MMP) are a specific group of enzymes that degrade the extracellular matrix through zinc-dependent proteolysis. MMP-9 plays a key role in podocyte injury, and podocyte migration and Adriamycin-induced cell injury may be related to the upregulation of MMP-9 expression. Sai et al. (18) found that the expression levels of MMP-2 and MMP-9 were decreased in a doxorubicin-induced podocyte injury group, while the use of Astragali Radix in the intervention group inhibited this decrease and prevented podocyte injury.
The GO analysis showed that target genes were mainly involved in biological processes, such as signal transduction, the positive regulation of cell proliferation, and the positive regulation of migration. The cellular components included the plasma membrane, extracellular exosome, and extracellular space. The molecular functions included protein binding, zinc-ion binding, protein tyrosine kinase activity, and enzyme binding. This suggests that Astragali Radix may regulate the generation and proliferation of podocytes and inhibit the apoptosis induced by oxidative stress in the treatment of NS.
The regulation of the cell membrane may be related to the regulation of lipids. Podocyte injury is considered one of the most critical factors in the development of proteinuria caused by glomerular filtration barrier dysfunction, and continuous injury may lead to severe apoptosis (19). Astragali Radix water extract (20) improves albuminuria in rats with Adriamycin nephropathy by inhibiting oxidative stress and endothelium-type nitric oxide synthase. Clinical manifestations of NS include high cholesterol, hyperlipidemia, and hypoproteinemia, which has been found to be related to lipid metabolism disorders (21). Liu et al. (22) found that Astragali Radix regulates lipid metabolism and improves the ability of anti-lipid peroxidation. This is consistent with the results of the present study. Considering the role of Astragali Radix in pathways in cancer and in diabetic complications, the application of Astragali Radix in various cancers and diabetes is also worth researching in the future.
Conclusions
In the present study, we used the network pharmacology method to explore the potential targets and pathways of Astragali Radix in the treatment of NS, and provided future research directions for the treatment of NS with a complex pathogenesis.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-348
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-348). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Agrawal S, Zaritsky JJ, Fornoni A, et al. Dyslipidaemia in nephrotic syndrome: mechanisms and treatment. Nat Rev Nephrol 2017;14:70. [Crossref] [PubMed]
- Bagga A, Mantan M. Nephrotic syndrome in children. Indian J Med Res 2005;122:13-28. [PubMed]
- Greenbaum LA, Benndorf R, Smoyer WE. Childhood nephrotic syndrome--current and future therapies. Nat Rev Nephrol 2012;8:445-58. [Crossref] [PubMed]
- Wang W, Chen C, Fu Q. Effect of fangji huangqi decoction on podocin expression in kidney tissue of adriamycin nephropathy rats. J Tra Chi Med 2014;42:57-9.
- Yan Q, Yan T, He B. Effects of different proportions of angelica tonifying blood decoction on the proliferation of mesangial cells in renal tissue of rats under high glucose. Hubei J Tra Chi Med 2015;37:11-3.
- He H, Wang J, Chen J. Research progress on pharmacological action of main active components of Astragalus membranaceus. Latset Med Infor 2016;16:49-50.
- Wang Y, Zhao Y, Mei C. Research progress of astragalus in clinical application. Latset Med Infor 2016;16:216-9.
- Zuo C, Xie XS, Qiu HY, et al. Astragalus mongholicus ameliorates renal fibrosis by modulating HGF and TGF-beta in rats with unilateral ureteral obstruction. J Zhejiang Univ Sci B 2009;10:380-90. [Crossref] [PubMed]
- Ren S, Qian Y, Liu Q. Effect of Astragalus membranaceus on podocytes in rats with adriamycin nephropathy. Chin J Neph Int Tra West Med 2009;10:851-3.
- Mei Z, Ding L, Shi T, et al. Effect of Astragalus membranaceus on oxidative stress in patients with nephrotic syndrome. Chin J Neph Int Tra West Med 2004;5:209-11.
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008;4:682-90. [Crossref] [PubMed]
- Liu Z, Liu H, Liu K, et al. Mechanism analysis of Naozhenning granules in the treatment of traumatic brain injury based on network pharmacology. Chin Herb Med 2018;49:3531-40.
- Dan W, Yao G, Huan X, et al. Study on anti-depression mechanism of Bupleurum based on network pharmacology. J Phar Sci 2018;53:210-9.
- Jing L, Li G, Yao G, et al. Study on targets of cough-relieving and phlegm-relieving active components of Scolander's flower based on network pharmacology. Chin Herb Med 2018;49:179-87.
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- von Mering C, Jensen LJ, Snel B, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005;33:D433-7. [Crossref] [PubMed]
- Li AP, Yang L, Cui T, et al. Uncovering the mechanism of Astragali Radix against nephrotic syndrome by intergrating lipidomics and network pharmacology. Phytomedicine 2020;77:153274 [Crossref] [PubMed]
- Sai YP, Song YC, Chen XX, et al. Protective effect of astragalosides from Radix Astragali on adriamycin-induced podocyte injury. Exp Ther Med 2018;15:4485-90. [Crossref] [PubMed]
- Pavenstädt H, Kriz W, Kretzler M. Cell biology of the glomerular podocyte. Physiol Rev 2003;83:253-307. [Crossref] [PubMed]
- You H, Lu Y, Gui D, et al. Aqueous extract of Astragali Radix ameliorates proteinuria in adriamycin nephropathy rats through inhibition of oxidative stress and endothelial nitric oxide synthase. J Ethnopharmacol 2011;134:176-82. [Crossref] [PubMed]
- Vaziri ND. Disorders of lipid metabolism in nephrotic syndrome: mechanisms and consequences. Kidney Int 2016;90:41-52. [Crossref] [PubMed]
- Liu C, Zheng X. Effect of astragalus on blood lipid and lipid peroxidation in hyperlipidemia mice. J Yanbian Uni Med 2006;18:178-80.



