
Polymorphisms in the interleukin 4 promoter -589C/T gene and the risk of asthma: a systematic review and meta-analysis
Introduction
Bronchial asthma (asthma for short) is a chronic inflammatory disease of the airways characterized by respiratory obstruction caused by bronchial hyperresponsiveness. It is jointly involved in a variety of cells and cytokines, and is related to the T helper type 2 (Th2) immune response (1). In the development of the occurrence of asthma, Th1/Th2 imbalance denotes allergic asthma is an important mechanism of the disease. Interleukin 4 (IL-4) is a key factor in inducing differentiation of Th2 cells (2), which is produced by Th2, reacts toTh2, and plays a pro-inflammatory role in asthma. The IL-4 gene, located on the long arm of chromosome 5, is about 10 kb, and is one of the larger lymphocytes. The gene consists of 4 exons and 3 introns. 5q21-33, 6q21-23, 11ql3, 12q14-24 are important susceptible regions of asthma on human chromosome, and many genes related to the occurrence and expression of asthma are distributed in these regions. Among them, IL-4 and IL-13 genes are the two candidate genes that have attracted much attention in relevant studies at home and abroad. They are located on the same chromosome (5q3l-q33, 5q23-31, respectively), so they are closely related to each other. Moreover, they have the generation of cytokines and share certain same parts in function and structure.
The mutation C→T at the -590 site of the IL-4 gene promoter is associated with the development of asthma, and the T allele can cause the increase of total IgE in plasma. At the same time, EOS activation, proliferation, release a variety of pro-inflammatory mediators, cause chronic allergic reactions, and destroy their own tissues and functions. A number of studies have suggested that IL-4 gene polymorphism is closely related to bronchial asthma and respiratory hyperresponsiveness (3), but there have been some differences in the conclusions of recent studies, which have also involved small sample sizes (4). A number of studies have suggested that high expression of the IL-4-590C/T gene may be an important cause of the inducement of bronchial asthma (5,6). Results of different studies are uncertain and inconsistent due to differences in research methods, sample size, region and race. A single study may not provide reliable evidence. Therefore, in order to further clarify the relationship between IL-4 gene polymorphism and bronchial asthma, we conducted a systematic review and meta-analysis of the existing available evidence. We present the following article in accordance with the PRISMA reporting checklist (available at https://dx.doi.org/10.21037/tp-21-419).
Methods
Search strategy
We systematically searched the databases of PubMed, Embase, Cochrane Library, China National Knowledge Infrastructure (CNKI), Wanfang, Chongqing VIP (CQVIP), and Chinese Biomedical Literature (CBM) from establishment of the database to April 2021. The following search terms were used: wheezing OR asthma. We checked the list of references to relevant articles to see if these references included other research reports that might be included in the review.
Literature inclusion and exclusion criteria
Articles were selected for inclusion if they met all the following criteria: (I) study type was randomized controlled trial (RCT) study, (II) language was limited to Chinese and English, (III) outcome of interest was wheezing/asthma (diagnosed by doctor or retrieved from Medicare database), (IV) measures of association [odds ratio (OR), hazard ratio (HR), or relative risk (RR)] and their 95% confidence interval (CI). If a study had several periods of follow-up, we used the values from the longest follow-up.
The exclusion criteria were as follows: repeated publication; research without full text, incomplete information or inability to conduct data extraction; animal experiments; reviews and systematic reviews. Unpublished or grey literature data (for example, in terms of conference abstracts, dissertations, or editorials) were not considered. The studies were selected by 2 reviewers independently, and any discrepancies were resolved by discussion, with adjudication from a third reviewer where necessary.
Literature screening and data extraction
The literature search, screening, and information extraction were completed independently by 2 researchers. In the event of doubts or disagreements, consensus was reached after discussion or consultation with a third party. The data extraction included the author, year, study type, number of cases, and outcome measures.
Literature quality assessment
Literature quality evaluation was independently conducted by 2 researchers, using the software Review Manager 5.3 (RevMan, Copenhagen: The Nordic Cochrane Center, The Cochrane Collaboration, 2014) risk assessment tool. According to the Cochrane risk assessment scale, allocation hiding, blinding, random sequence generation, whether the research results were blindly evaluated, and the result data were complete, the included studies were evaluated based on gender, choice of research report results, other biases, and so on. Any disagreements between researchers were reconciled through discussion or consultation with a third party. This meta-analysis was performed based on the related items of the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement.
Data synthesis and statistical analysis
The software RevMan 5.3 was used to analyze the data. We used RR (95% CI) as the binary variable, and standardized mean difference [SMD (95% CI)] combined effect size was used as the continuous variable. Heterogeneity was evaluated using I2. If the heterogeneity test revealed P≥0.1 and I2≤50%, homogeneity between studies was inferred, and the fixed effects model is used for combined analysis; if P<0.1 and I2>50%, it indicated heterogeneity between studies, and sensitivity analysis or subgroup analysis was conducted to investigate the source of heterogeneity. If the heterogeneity was still large, the random effects model was used, or combined results were relinquished and descriptive analysis was used instead. A funnel plot was used to analyze publication bias.
Results
Literature search results
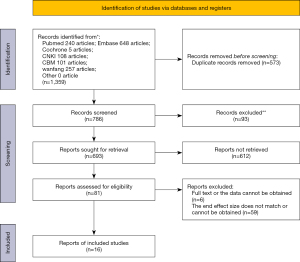
A total of 1,359 citations were identified: 240 from PubMed, 648 from Embase, 5 from Cochrane, 108 from CNKI, 101 from CBM, and 257 from Wanfang. Of these, 573 were excluded as duplicates and 93 were excluded after the first screening based on abstracts or titles, leaving 693 articles for full-text review. A further 612 were excluded that did not match the research content by reading the abstract, leaving 81 articles for screening based on title and abstract. A total of 59 articles that did not conform to the end point effect size or could not deliver the effect size data and experimental design were excluded after reading of full texts, leaving 22 full articles for review. An additional 6 were excluded due to unavailability of full text and literature data, leaving 16 articles for final inclusion in meta-analysis in Figure 1.
Baseline characteristics and quality assessment of the included studies
Baseline characteristics
The baseline characteristics and quality assessment of the included studies are shown in Table 1.
Table 1
| Author | Year | Selection | Comparability | Exposure | Score |
|---|---|---|---|---|---|
| Cao |
2012 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Liang |
2014 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Wang |
2012 | ☆☆ | ☆☆ | ☆☆ | 6 |
| Chiang |
2007 | ☆☆☆ | ☆☆ | ☆☆ | 7 |
| Sandford |
2000 | ☆☆☆ | ☆☆ | ☆☆ | 7 |
| Mak |
2007 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Narożna |
2016 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Zhorina |
2019 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Li |
2016 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Dahmani |
2016 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Kabesch |
2006 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Wu |
2010 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Daneshmandi |
2012 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Huang |
2012 | ☆☆☆ | ☆☆ | ☆☆☆ | 8 |
| Micheal |
2013 | ☆☆ | ☆☆ | ☆☆☆ | 7 |
| Li |
2014 | ☆☆☆ | ☆☆ | ☆☆ | 7 |
The offset risk of 9 references in this study was evaluated according to the NOS. The basic score was 7 stars or above, and only one was 6 points, so the overall quality was high. NOS, Newcastle-Ottawa Scale.
Quality assessment of the included studies
The quality assessment of the included studies was as follows:
Results of meta-analysis
A total of 1,359 citations were identified: 240 from PubMed, 648 from Embase, 5 from Cochrane, 108 from CNKI, 101 from CBM, and 257 from Wanfang. Of these, 786 were excluded as duplicate; 93 were excluded for being systematic evaluations, reviews, and animal studies; 612 were excluded for containing inconsistent research content as detected by reading the abstract. A further 59 were excluded for literature that did not conform to the endpoint effect size or the inability to obtain the effect size data and did not conform to the experimental design by reading the full text, leaving 22 articles for full-text review. An additional 6 articles were excluded due to the unavailability of full text and literature data. A total of 16 references were included in the meta-analysis.
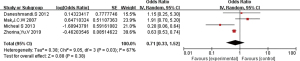
Meta-analysis of the effect of CT, CC, TT, C, and T polymorphism on asthma
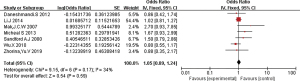
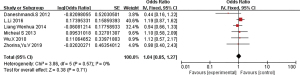
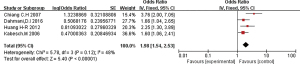
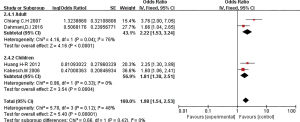
The 16 included literatures were combined and analyzed by RevMan 5.5.3 software, the results were as follows: CT, CC, and T gene polymorphisms were risk factors for asthma (OR =1.05, 95% CI: 0.89–1.24; OR =1.04, 95% CI: 0.85–1.27; OR =1.98, 95% CI: 1.54–2.53, respectively) (Figures 2-4). The TT and C gene polymorphisms were not risk factors for asthma (OR =0.71, 95% CI: 0.33–1.52; OR =0.95, 95% CI: 0.76–1.19) (Figures 5,6). The funnel plot is shown in Figure 6. It can be seen that the funnel plot is mainly meristic, this indicate that there was no obvious publication bias in this study.

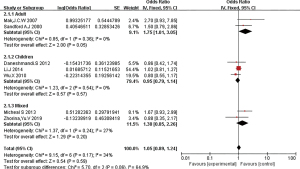
Subgroup analysis
CT subgroups
Ethnic subgroup analysis showed that genotype CT was associated with the risk of asthma in the Asian population (OR =1.75, 95% CI: 1.01–3.05). However, the OR in the children group was 0.95 (95% CI: 0.79–1.14), indicating that CT gene polymorphism was not a risk factor for asthma. In the mixed group, the OR =1.38 (95% CI: 0.85–2.26), indicating that CT gene polymorphism is a risk factor for asthma (Figure 7).
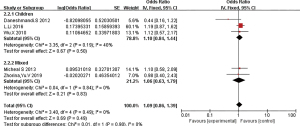
CC subgroups
Ethnic subgroup analysis showed that genotype CC was associated with the risk of asthma in the Asian population (OR =1.09, 95% CI: 0.86–1.39). The OR was 1.1 (95% CI: 0.84–1.44) and 1.06 (95% CI: 0.63–1.79) in the children group and the mixed group, respectively, indicating that CC gene polymorphism is a risk factor for asthma (Figure 8).
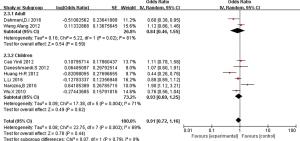
C subgroups
Ethnic subgroup analysis showed that genotype C was not associated with the risk of asthma in the Asian population (OR =0.91, 95% CI: 0.72–1.16). The OR was 0.93 (95% CI: 0.69–1.25) and 0.84 (95% CI: 0.46–1.55) in the children group and the adult group, respectively, indicating that C gene polymorphism was not a risk factor for asthma (Figure 9).
T subgroups
Ethnic subgroup analysis showed that genotype C was associated with the risk of asthma in the Asian population (OR =1.98, 95% CI: 1.54–2.53). The OR was 2.22 (95% CI: 1.53–3.24) and 1.81 (95% CI: 1.3–2.51) in the adult group and the children group, respectively, indicating that T gene polymorphism was a risk factor for asthma (Figure 10).
Publication bias
The funnel plot is shown in Figure 11. It can be seen that the funnel plot is mainly meristic, this indicate that there was no significant publication bias in this study.
Sensitivity analysis
Sensitivity analysis was conducted by eliminating each included study individually, and conducting a summary analysis of the remaining studies to assess whether any single included study had an extreme impact on the results of the whole meta-analysis. None of the studies had a significant impact on the results of the meta-analysis, pointing that the results of the other studies were steady and reliable.
Discussion
Mechanisms of asthma
Asthma is a respiratory disease characterized by recurrent airway obstruction, respiratory hyper responsiveness, and chronic bronchial inflammation. Adult asthma is more likely to develop into persistent asthma, the severity of such asthma is gradually worsening, and the prognosis is poor. The severity of asthma is an important factor affecting the number of acute attacks, duration of symptoms, and the treatment effect of patients (21-23). One of the core factors of asthma attack is the high activation of Th2, and IL-4 plays an important role as the key component of Th2 activation. The function of IL-4 is related to multiple factors, among which the -589C/T polymorphism has been widely studied (24).
Relationship between IL-4 gene polymorphism and asthma
The main reasons for the increased risk of asthma caused by the mutation of the IL-4 gene promoter site are as follows: the mutation of the base in the IL-4 gene promoter region may increase the production of IL-4 in human body, thus leading to the occurrence of asthma; Secondly, IL-4 has many roles in the body, mainly producing a variety of biological activities in the human body, so it plays a very important role in the occurrence and development of asthma. When the body is stimulated by inflammation, IL-4 binds to its receptor to promote the differentiation of Th0 into Th2 cells and produce a large number of cytokines. On the other hand, it prevents the transformation of Th0 into Th1 and inhibits the function of Th1. In addition, IL-4 can also promote the expression of MHCII class antigen CD40 in B lymphocytes, so that B cells proliferate and produce more IgE. At the same time, the expression of CD23 and FcεRI amplifies and enhances the production and effect of IgE. IL-4 also prevents the death of B cells and enhances the expression of human IgE Fc receptor II by B cells and the transcription of FCCR mRNA, promoting the synthesis of SFCCR Cin, and thereby elevating the IgE level in the body. In addition, IL-4 can also increase the proliferation of vascular endothelium and increase the expression of vascular cell adhesion molecule 21 (VCAM21) by endothelial cells, which plays a role in the development of asthma.
Researchers have found a number of polymorphisms in the IL-4 gene that are associated with a variety of asthma phenotypes. Wang et al. (26) found that gene polymorphism at locus 589 in the promoter region of IL-4 gene can affect the degree of airway obstruction in asthmatic patients and may be related to the severity of asthma attack. Sandford et al. (10) also showed that the 589T allele was associated with asthma, while Hijazi (27) found that the C590T polymorphism of IL-4 gene was weakly correlated with the incidence of asthma.
Principal findings
It was found that CT, TT, and T gene polymorphisms were risk factors of asthma, and CC and T gene polymorphisms were risk factors of asthma by subgroup analysis. This meta-analysis on 16 studies provided considerable evidence indicating that IL-4-589C/T gene -589C/T polymorphism is associated with the risk of asthma. Nie et al. (25) investigate the association between polymorphism in the IL-4 and asthma susceptibility fond that the IL-4 -589C/T polymorphism was a risk factor of asthma. Compared with the findings of Nie et al., the susceptibility of asthma with IL-4-589C/T polymorphism in adults, children and mixed population was studied in this study, and the grouping was more reasonable. The researchers found a number of polymorphisms in the IL-4 gene that are associated with a variety of asthma phenotypes. Wang et al. (26) found that gene polymorphism at locus 589 in the promoter region of IL-4 gene can affect the degree of airway obstruction in asthmatic patients, and may be related to the severity of asthma attack. Sandford et al. (10) also showed that the 589T allele was associated with asthma, while Hijazi et al. (27) found that the C590T polymorphism of IL-4 gene was weakly correlated with the incidence of asthma. However, Cui et al. found no correlation between IL-4-589C/T and allergic asthma in Chinese people (28). The reason for this difference may be due to heterogeneity of races and regions, the distribution of gene polymorphisms may also vary greatly. A subgroup analysis was conducted, and it was found that this locus had a certain correlation with asthma. However, as this study was only conducted between children and adults, and there was a small amount of literature, further study is needed to determine whether the study results are generalizable to the whole population.
IL-4-589C/T polymorphism and other diseases
Sivangala et al. through the study of cytokine gene polymorphisms in patients with tuberculosis and their household contacts found that IL-4 (-589C/T) gene polymorphisms may be associated with TB susceptibility (29). However, in the study on the correlation between Graves’ hyperthyroidism and IL-4 promoter (-590C/T) gene polymorphism, Lv et al. (30) found that there was no correlation between GD hyperthyroidism and IL-4 (-590) gene polymorphism in Tianjin Han population. Also Dai et al. (31) on IL-4 gene promoter -590C>T was found in the study of the correlation between T polymorphism and HCV chronic infection in Yunnan Han population. T polymorphism has no correlation with HCV chronic infection in Yunnan Han population.
Limitations
There were some noteworthy limitations to this study: (I) it was not possible to analyze gene-gene and gene-environment interactions; (II) there were differences in age and gender among literatures; (III) the number of included literatures in this study was small, and different regions used different detection methods in the process of sample collection; (IV) the language of literature was limited to Chinese and English, and articles in other languages and unpublished articles were not included. The above limitations may have led to bias of the results to some extent.
Conclusions
In conclusion, through the study of IL-4(C-590T) gene polymorphism, it was found that CT, TT, and T gene polymorphism were risk factors for asthma, and the results suggested that this locus polymorphism was related to the risk of asthma. The results of subgroup analysis showed that CC and T gene polymorphisms were risk factors for asthma. Thus, IL-4 (C-590T) may be the susceptibility gene of asthma. Further functional studies with large samples and multiple loci are needed to investigate whether the IL-4 gene plays a role in susceptibility to asthma.
Acknowledgments
Funding: This study was supported by Sichuan Province Birth Defects Clinical Medical Research Center Project (2019YFS0531-2).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-419
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-419). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Falcai A, Soeiro-Pereira PV, Kubo CA, et al. Peripheral blood mononuclear cells from severe asthmatic children release lower amounts of IL-12 and IL-4 after LPS stimulation. Allergol Immunopathol (Madr) 2015;43:482-6. [Crossref] [PubMed]
- Zhang JH, Zhang M, Wang YN, et al. Correlation between IL-4 and IL-13 gene polymorphisms and asthma in Uygur children in Xinjiang. Exp Ther Med 2019;17:1374-82. [PubMed]
- Chiang CH, Tang YC, Lin MW, et al. Association between the IL-4 promoter polymorphisms and asthma or severity of hyperresponsiveness in Taiwanese. Respirology 2007;12:42-8. [Crossref] [PubMed]
- Zhang JH, Zhou GH, Wei TT, et al. Association between the interleukin 4 gene -590C>T promoter polymorphism and asthma in Xinjiang Uighur children. Genet Mol Res 2016;15: [Crossref] [PubMed]
- Zheng S, Zhu X, Li B, et al. Correlation of gene polymorphism of interleukin 4 receptor alpha peptide chain and total serum IgE levels in asthmatic children in Guiyang area. Zhonghua Yi Xue Za Zhi 2014;94:2822-7. [PubMed]
- Huang HR, Zhong YQ, Wu JF. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene 2012;494:96-101. [Crossref] [PubMed]
- Cao Y, Cui Q, Tang C, et al. The association of IL-4 and IL-13 gene polymorphism with asthma in children. Chin J Modem Med 2012;22:31-5.
- Liang W, Zhou Z, Ji Z, et al. Association study of bronchial asthma with polymorphisms of IL-4 and IL-4R receptor genes. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2014;31:97-100. [PubMed]
- Wang A, Qian P, Zhou L, et al. The relationship between single nucleotide polymorphisms of IL-4 and IL-13 and asthma in Chinese Han population. Chin J Asthma 2012;6:420-4. (Electronic Edition).
- Sandford AJ, Chagani T, Zhu S, et al. Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol 2000;106:135-40. [Crossref] [PubMed]
- Mak JC, Ko FW, Chu CM, et al. Polymorphisms in the IL-4, IL-4 receptor alpha chain, TNF-alpha, and lymphotoxin-alpha genes and risk of asthma in Hong Kong Chinese adults. Int Arch Allergy Immunol 2007;144:114-22. [Crossref] [PubMed]
- Narożna B, Hoffmann A, Sobkowiak P, et al. Polymorphisms in the interleukin 4, interleukin 4 receptor and interleukin 13 genes and allergic phenotype: A case control study. Adv Med Sci 2016;61:40-5. [Crossref] [PubMed]
- Zhorina YV, Abramovskikh OS, Ignatova GL, et al. Analysis of associations of polymorphisms in the genes coding for L4, IL10, IL13 with the development of atopic bronchial asthma and its remission. Bulletin of RSMU 2019;87-91. [Crossref]
- Li L, Li Y, Zeng XC, et al. Role of interleukin-4 genetic polymorphisms and environmental factors in the risk of asthma in children. Genet Mol Res 2016;15: [Crossref] [PubMed]
- Dahmani DI, Sifi K, Salem I, et al. The C-589T IL-4 Single Nucleotide Polymorphism as a Genetic Factor for Atopic Asthma, Eczema and Allergic Rhinitis in an Eastern Algerian Population. Int J Pharm Sci Rev Res 2016;37:213-23.
- Kabesch M, Schedel M, Carr D, et al. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol 2006;117:269-74. [Crossref] [PubMed]
- Wu X, Li Y, Chen Q, et al. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle china. J Asthma 2010;47:238-44. [Crossref] [PubMed]
- Daneshmandi S, Pourfathollah AA, Pourpak Z, et al. Cytokine gene polymorphism and asthma susceptibility, progress and control level. Mol Biol Rep 2012;39:1845-53. [Crossref] [PubMed]
- Micheal S, Minhas K, Ishaque M, et al. IL-4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients. J Investig Allergol Clin Immunol 2013;23:107-11. [PubMed]
- Li J, Lin LH, Wang J, et al. Interleukin-4 and interleukin-13 pathway genetics affect disease susceptibility, serum immunoglobulin E levels, and gene expression in asthma. Ann Allergy Asthma Immunol 2014;113:173-179.e1. [Crossref] [PubMed]
- Erdoğan Yüce G, Taşcı S. Effect of pranayama breathing technique on asthma control, pulmonary function, and quality of life: A single-blind, randomized, controlled trial. Complement Ther Clin Pract 2020;38:101081 [Crossref] [PubMed]
- Prem V, Sahoo RC, Adhikari P. Comparison of the effects of Buteyko and pranayama breathing techniques on quality of life in patients with asthma - a randomized controlled trial. Clin Rehabil 2013;27:133-41. [Crossref] [PubMed]
- Sankar J, Das RR. Asthma - A Disease of How We Breathe: Role of Breathing Exercises and Pranayam. Indian J Pediatr 2018;85:905-10. [Crossref] [PubMed]
- Hussein IA, Jaber SH. Genotyping of IL-4 -590 (C>T) Gene in Iraqi Asthma Patients. Dis Markers 2017;2017:5806236 [Crossref] [PubMed]
- Nie W, Zhu Z, Pan X, Xiu Q. The interleukin-4 -589C/T polymorphism and the risk of asthma: a meta-analysis including 7,345 cases and 7,819 controls. Gene 2013;520:22-9. [Crossref] [PubMed]
- Wang W, Halmurat W, Yilihamu S, et al. A study on the relationship between interleukin-4 promoter polymorphism and asthma in a Xinjiang Uyger population. Zhonghua Jie He He Hu Xi Za Zhi 2004;27:460-4. [PubMed]
- Hijazi Z, Haider MZ. Interleukin-4 gene promoter polymorphism C590T and asthma in Kuwaiti Arabs. Int Arch Allergy Immunol 2000;122:190-4. [Crossref] [PubMed]
- Cui T, Wu J, Pan S, et al. Polymorphisms in the IL-4 and IL-4R [alpha] genes and allergic asthma. Clin Chem Lab Med 2003;41:888-92. [Crossref] [PubMed]
- Sivangala R, Ponnana M, Thada S, et al. Association of cytokine gene polymorphisms in patients with tuberculosis and their household contacts. Scand J Immunol 2014;79:197-205. [Crossref] [PubMed]
- Lv YJ. Correlation between hyperthyroidism in Graves disease and il-4 promoter (-590C/T) gene polymorphism and serum IL-4 and IL-2 levels after glucocorticoid treatment. Tianjin: Tianjin Medical University, 2005.
- Dai SY, Dai SP, Yu Z, et al. Association of SNP-1098T>G and-590C>T in IL-4 Gene Promoter with Chronic HCV Infection in Yunnan Han Population. Journal of Kunming Medical University 2015;36:9-12.
(English Language Editor: J. Jones)



