
Compound heterozygous pathogenic variants in the GALC gene cause infant-onset Krabbe disease
Introduction
Krabbe disease is a rare, autosomal recessive, metabolic and neurodegenerative disease with an estimated incidence of 1 in every 100,000 live births. This disorder affects the white matter of the central and peripheral nervous systems and is therefore also called globoid cell leukodystrophy (1). It is caused by deficiency of the lysosomal protein, galactosylceramidase (GALC), which results in the impaired degradation of a major myelin lipid, galactocerebroside (2).
Krabbe disease varies in its age of onset and clinical course, and its subtypes include infantile type (onset before 12 months of age), late-infantile type (onset between 12 and 36 months), juvenile type (onset between 3 to 16 years), and adult type (onset over 16 years) (3,4). Infantile-onset disease mainly manifests as extreme irritability, spasticity, feeding difficulties, and regression of psychomotor development. However, for late-onset cases, the disease course and presentation of clinical features vary and can include muscle weakness, vision loss, arrest of intellectual development, and ataxia (5). For example, juvenile onset, occurring between 3 and 16 years of life, is typified by cognitive regression, abnormal behaviors, and ataxia. While blindness is often described as a characteristic finding of the disease, it is more common in the infantile phenotype, where vision loss typically arises in the late stages of disease. In comparison, reports of vision loss in late onset phenotypes are less well-described and may be subject to variation between genotypes (6).
Krabbe disease is usually diagnosed after a comprehensive consideration of clinical features, neuroimaging results, and electrophysiological findings. Further, the diagnosis is established by the detection of deficient GALC enzyme activity in leukocytes or elevated psychosine levels. Thus far, the standard treatment for Krabbe disease has been hematopoietic stem cell transplantation (HSCT) for both early- and late-onset disease (7-9). Pathogenic GALC gene variants can confirm abnormal enzyme activity and provide solid evidence for diagnosis (10). Although 471 variants of the GALC gene have been reported in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar), only 160 are categorized as pathogenic or likely pathogenic, the presence of which is crucial for clinical diagnosis. However, 179 variants have been reported to have conflicting or uncertain significance and require clinical and experimental validation to clarify their pathogenicity.
In Krabbe disease, mutations in GALC cause psychosine accumulation, inducing demyelination, a neuroinflammatory “globoid” reaction and neurodegeneration. Using a novel Krabbe disease model, Weinstock et al. (11) showed Galc-deficient Schwann cells autonomously produce psychosine. Besides, Galc-deficient macrophages were transformed into globoid cells by exposure to galactosylceramide (GALC substrate) and produce a more severe Krabbe disease phenotype. Thus, Krabbe disease may be caused by at least two mechanisms: psychosine-induced demyelination and secondary neuroinflammation from galactosylceramide storage in macrophages.
In this study, we introduced compound heterozygous variants and comprehensively investigated the clinical features, enzyme activity, genetic testing, and molecular function validation of GALC gene variants in infant-onset Krabbe disease.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-403).
Methods
Clinical examination
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the Third Affiliated Hospital of Zhengzhou University (No. 32, 2019) and informed consent was taken from parents/guardian of the patient. Clinical information collection, routine blood examination, electromyogram (EMG), and magnetic resonance imaging (MRI) were performed, and query into the family history of genetic disorders was made.
Whole exome sequencing
After family consent was given, blood samples were drawn from the patient and his parents. Whole exome sequencing was then outsourced to Ciphergene (Beijing, China). Database filtering and variant interpretation were subsequently performed to identify any potential variants that could be driving the disease.
Enzyme activity detection
GALC activity in patient’s leukocytes was analyzed with the use of 6-hexadecanoylamino-4-methylumbelliferyl-β-D-galactoside (HMU-βGal) as a fluorogenic substrate in the Clinical Laboratory of Zhongke, Beijing. The galactosidase can hydrolyze HMU-βGal to produce specific fluorescence products under acidic conditions. The fluorescence value was measured by a F97 series fluorescence spectrophotometer at 365 and 460 nm (12-14). According to the standard curve and the specific fluorescence value, the activity of GALC enzyme can be calculated. Enzyme activity is expressed as nmol/g/min protein.
Validation of variants from blood
Total RNA was extracted from the patient’s peripheral blood sample using the RNAiso Plus kit (108/9109, TaKaRa, Shiga, Japan), and complement DNA (cDNA) was transcribed using the PrimeScript RT reagent Kit with gDNA Eraser (RR047A, TaKaRa). Polymerase chain reaction (PCR) was performed and evaluated on a 1% agarose gel.
Alternative splicing validation by minigene
Genomic DNA was extracted from the patient’s peripheral blood, and the following two-pair nested PCR primers were designed for target fragment amplification: 4181-GALC-F (5'-AGTTATTCTTAATAGTAACA-3'), 7339-GALC-R (5'-GTACACATTCTCACAGAACT-3'), 4459-GALC-F (5'-CAGCCCTACTTGCCCAATGA-3'); and 6998-GALC-R (5'-GCATCATAGGAAACATGGAA-3'). Expression primers were designed as follows: pcDNA3.1-GALC-BamHI-F (5'-GCTCGGATCCATGGCAACCTCCCGACTTCTAGT-3') and pcDNA3.1-GALC-XhoI-R (5'-TAGACTCGAGCCCAATGAGTGTAATATTGGGA-3'). After amplification, enzyme digestion, and ligation, variant and wild-type minigenes of GALC were cloned into the pcDNA3.1 vector. pcDNA3.1-GALC-wt/mut was transfected into the 293T cell line. Cells were harvested after 48 hours for further validation of alternative splicing. Total RNA was extracted from 293T cells using the RNAiso Plus kit (108/9109, TaKaRa) according to the manufacturer’s instructions. First-strand cDNA was synthesized using a Prime Script RT Reagent kit with gDNA Eraser (RR047A, TaKaRa). PCR was performed and evaluated on a 1% agarose gel.
GALC overexpression plasmid construction
Total RNA was extracted from blood samples using the RNAiso Plus kit (108/9109, TaKaRa) and reverse-transcribed to cDNA as a template for PCR to construct the GALC expression plasmid. Full-length open reading frame sequences were amplified from the cDNA using PCR. The PCR product and vector were digested by BamHI and XhoI. After ligation and sequencing validation, the target fragment was subcloned into the restriction sites of the pHAGE vector.
Cell culture and transient transfection
The experiments were performed using the 293T cell line (GDC0187, CCTCC). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum. Wild-type and variant plasmids were transfected into 293T cells using the Lipofectamine 2000 Transfection Reagent (11668019, Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Briefly, the vector DNA and Lipofectamine 2000 reagent mixture was prepared by dilution in Opti MEM medium (Gibco, Grand Island, NY, USA), followed by thorough mixing. The mixture was incubated for 15 minutes at room temperature before the DNA-lipid complex was added to 293T cells.
Real-time PCR (RT-PCR)
Total RNA was extracted from 293T cells using the RNAiso Plus kit (108/9109, TaKaRa) according to the manufacturer’s protocol. First-strand cDNA was synthesized using a Prime Script RT Reagent kit with gDNA Eraser (RR047A, TaKaRa). Quantitative RT-PCR analysis was performed using SYBR Green Realtime PCR Master Mix (QPK-201, Toyobo, Osaka, Japan) in a CFX Connect Real-Time PCR System (Bio-Rad, Hercules, CA, USA). GALC messenger RNA (mRNA) abundance was measured by quantitative PCR. The data were analyzed using the standard curve method and the geometric mean of the internal control gene, ACTB.
Western blotting
Protein was extracted from transfected cells using the RIPA lysis and extraction buffer (C1053, PPLYGEN, Beijing, China). Protein concentration was measured using a bicinchoninic acid (BCA) assay kit (20201ES76, Yeasen Biotechnology, Shanghai, China). Total protein (10 µg/lane) was loaded onto SDS-PAGE and transferred to a polyvinylidene difluoride membrane. After being blocked with 5% skim milk for 1 hour, the membranes were incubated with the primary antibody at 4 °C overnight. The primary antibodies used in this study were mouse polyclonal anti-HA tag (1:2,000, 2063, QB, Wuhan, China) and anti-tubulin (1:1,500, ABclonal Technology, Woburn, MA, USA).
Statistical analysis
Statistical two-tailed unpaired t-tests were performed with a confidence interval of 95%.
Results
Case presentation
The patient was the first child of a nonconsanguineous Chinese couple with no family history of genetic disorders. He was born by spontaneous delivery at 40 weeks and weighed 3,600 g. He developed pneumonia at 6 months of age and was hospitalized in the pediatric ICU for nearly a month. Developmental regression was observed at 7 months of age, at which point he could no longer sit alone or hold up his head. Two weeks later, he developed feeding difficulties, irritability, and decreased voluntary movements. He was admitted to our hospital at 8 months of age. Physical examination revealed a normal head circumference of 45 cm, but the patient demonstrated an inability to grasp with his hands and support his trunk and lower limbs. Physical examination also showed increased muscle tonus in all limbs.
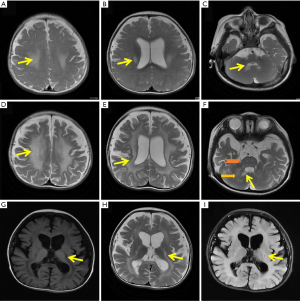
Gas chromatography of organic acids in urine and tandem mass spectrometry through acylcarnitine profiles analysis in dry blood filter paper were normal. Other test results, such as serum levels of homocysteine, lactic acid, and blood ammonia, were also in the normal range. Limb EMG revealed a generalized peripheral neuropathy. Additionally, MRI at 8 months revealed a combination of cerebral white matter changes in the central region, lateral ventricle, and cerebellar dentate nucleus (Figure 1A-1C). Re-examination at 14 months demonstrated a progressive course, including the anterior and posterior spread of demyelination, global atrophy, diffusely affected white matter, cystic degeneration of the pyramidal tract, and brainstem white matter changes (Figure 1D-1I).
Whole exome sequencing
Considering the clinical manifestations and findings of cranial MRI and electromyography, we focused our differential diagnosis on potential hereditary leukoencephalopathy. After family consent was obtained, blood samples were drawn from the patient and his parents. After sequencing, database filtering, and variant interpretation, whole exome sequencing revealed compound heterozygous variants, c.658C>T (p.Arg220*) and c.328+5G>T, in the GALC genes that were inherited from the mother and father, respectively. The c.658C>T mutant was found in the ClinVar database where it had been reported as pathogenic or likely pathogenic (https://www.ncbi.nlm.nih.gov/clinvar/variation/189024/). On the other hand, the c.328+5G>T mutation had not been reported in a public database (gnomAD or Exome Aggregation Consortium) and was designated as a variant of unknown significance (VUS) by the American College of Medical Genetics and Genomics (ACMG) guidelines (Table 1).
Table 1
| Gene | Variant | Inheritance | MAF | SIFT | Polyphen2 | MutationTaster | Evidence | Category | ||
|---|---|---|---|---|---|---|---|---|---|---|
| ExAc | gnomAD | 1000genome | ||||||||
| GALC | c.658C>T (p.Arg220*) | Mother | 8.36E-06 | 4.07E-06 | NE | – | – | A | PVS1 + PM2 | LP |
| c.328+5G>T | Father | NE | NE | NE | – | – | – | PM2 + PM3 + PP3 | VUS | |
Transcript, NM_ 000153.4. *, nonsense variant. MAF, minor allele frequency; ExAC, the exome aggregation consortium; gnomAD, the Genome Aggregation Database; NE, nonexistent; SIFT, sorting intolerant from tolerant; PVS1, pathogenic very strong; PM2/PM3, pathogenic moderate; PP3, pathogenic supporting; LP, likely pathogenic; VUS, variant of unknown significance.
Enzyme activity detection for galactocerebroside
The GALC variants implied the possibility of Krabbe disease, which is caused by GALC gene variants and is characterized by galactocerebroside deficiency in the serum, leukocytes, and fibroblasts. Thus, we measured the enzyme activity of galactocerebroside and observed significantly decreased galactocerebroside activity (0 nmol/g/min at month 8, 0.7 nmol/g/min at month 11; ref: 17.2–56.1).
Variants confirmed from blood
Both variants in the patient were observed in each parent: the heterozygous c.658C>T was detected in his mother, and the heterozygous c.328+5G>T was detected in his father (Figure 2A). A shorter band from the RT-PCR results demonstrated that alternative splicing occurred in the patient and his father (Figure 2B). The schematic map and sequence are shown in Figure 2C,2D.
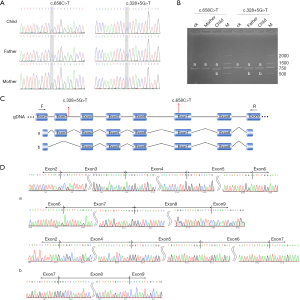
Variants result in abnormal RNA isoform and truncated protein
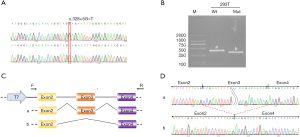
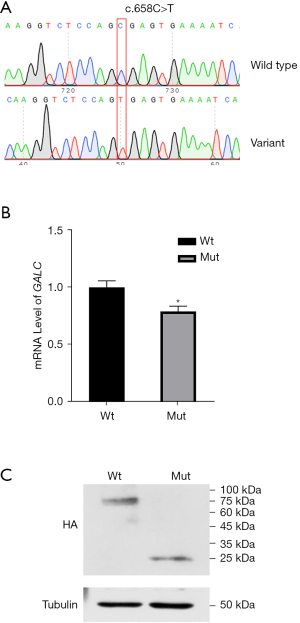
To further explore the effect of the c.328+5G>T variant on splicing, we cloned the variant and its wild type into the pcDNA3.1 vector. After confirmation by Sanger sequencing (Figure 3A), a shorter PCR band (Band B, Figure 3B) was observed in the 293T cell line. These results indicated alternative splicing resulting from the c.328+5G>T variant. The schematic map and sequence are shown in Figure 3C,3D. To evaluate the effect of the c.658C>T variant, we cloned the variant and its wild type into the pHAGE vector. After confirmation by Sanger sequencing (Figure 4A), quantitative RT-PCR was performed. Interestingly, a slight downregulation of RNA was observed (Figure 4B). Additionally, western blotting revealed a shorter band (~25.41 kDa) in the variant sample (Figure 4C), and the truncated protein indicated an early termination resulting from c.658C>T.
Discussion
The infantile type of Krabbe disease accounts for 85–90% of cases, and its main clinical features are rapid progression and high mortality. Children with this disease exhibit no symptoms in the first few months of life and gradually exhibit one or more of the following clinical symptoms: irritability, sensitivity to their environment, episodic fever of unknown etiology, epilepsy, psychomotor regression, and possible stiffness (3,4). For late-infantile, juvenile, and adult types, the clinical features include cognitive developmental delays, spastic tetraparesis, optic atrophy, and cerebellar ataxia. Typical MRI features include abnormal white matter, symmetric cerebral and cerebellar demyelination, and calcifications in the basal nuclei, corpus callosum, and intracranial sites (15). Moreover, dilatation in the subarachnoid spaces and ventricles or brain atrophy may be observed, especially in the late stage of Krabbe disease (16).
Large fragment deletions (exons 11–17, IVS10del30kb) are common variants in GALC (17); however, an increasing number of single-nucleotide polymorphism (SNP) variants have been found due to the rapid development of sequencing technology (18). We searched for variants in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar) and found that among the 471 variants collected, 160 were classified as pathogenic or likely pathogenic. Unlike large fragment deletions, defining the influence of SNPs on proteins is more challenging. More than 179 variants have been reported to have conflicting or uncertain significance, and additional clinical cases or experimental validation is required to clarify their pathogenicity. Altered enzymatic activity of galactocerebrosidase is crucial for the diagnosis of Krabbe disease, and the lack of enzyme activity can be observed in most cases identified as carrying pathogenic or likely pathogenic variants (https://www.ncbi.nlm.nih.gov/clinvar). The variant c.658C>T (p.Arg220*) was detected in our study, suggesting its function as a nonsense variant, and it was evaluated as likely being pathogenic. This variant was found in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/189024/) where it was reported to be pathogenic or pathogenic in other unrelated cases (19,20). Moreover, nonsense variants usually function in either nonsense-mediated mRNA decay or contribute to premature protein translation termination (truncated protein) (21). In our study, we observed this variant interfering with protein translation (Figure 4C).
Previous studies have revealed that several exon variants have a degree of function in RNA splicing; however, an increasing amount of evidence has been reported regarding the influence of variants in introns on RNA splicing (22), either through exon junction (23) or poison exon insertion (24). The c.328+5G>T variant found in our study influenced splicing and was located in intron regions of GALC between exons 3 and 4 (Figure 3C). Up to now, four cases (including our case) has been reported (Table 2), suggesting that variants in introns also play an important role on effecting the proteins.
Table 2
| Publication | Zygosity | Allele 1 | Allele 2 | Age of onset | Origin | Phenotype | Initial signs | Vison impairment |
|---|---|---|---|---|---|---|---|---|
| Zhao et al., 2018 | Hetero | c.1901T>C_p.L634S | c.195+1 G>A | 8 years 10 months | Chinese | Juvenile | Vision impairment | Yes |
| Tappino et al., 2010 | Hetero | c.1901T>C_p.L634S | c.1489+1G>A | 7 months | NA | Infantile | Spasticity | No |
| Furuya et al., 1997 | Hetero | c.1901T>C_p.L634S | c.582+5G>A | ~20 years | Japanese | Adult | Spastic paresis | No |
| Our case | Hetero | c.658C>T p.Arg220* | c.328+5G>T | ~6 months | Chinese | Infantile | Psychomotor regression | No |
*, nonsense variant.
Interestingly, another variant similar to the variant c.328+6A>G was found in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/314757/evidence/) and was classified as having unknown significance, in that the evidence for enzyme activity or clinical diagnosis of infantile Krabbe was absent. We validated the c.328+5A>G mutation in our sample, which revealed an exon junction splicing event (Figures 2B,2C,3B). We searched for variants in the introns of GALC: 80 variants were found, 19 of which were classified as pathogenic or likely pathogenic. Among these, 18 variants were categorized as being related to enzymatic activity deficiency, while 1 lacked any evidence. Unfortunately, none of these have been experimentally validated.
Bascou et al. reported a cohort with onset between 6 months and 3 years, they found median onset age was 11.5 months, with a median delay of 3.5 months between onset of symptoms and diagnosis. Among all 32 symptomatic patients, 23 onset before 12 months which was regards as infantile Krabbe disease. The most common initial signs and symptoms were loss of acquired developmental milestones, irritability, abnormal gait, motor delay, and abnormal muscle tone (4). Like reported cases, our case onset at 6 months, also presented irritability, abnormal muscle tone. However, our case presented a severe months development delay when evaluating at 8 month, around 5 months, which was larger than reported 3.5 months.
Early infantile Krabbe disease is a rapidly progressive and fatal disease, and HSCT is currently the only clinically available treatment option which can improve outcomes if performed prior to the onset of severe symptoms. Although there is some relationship between genetic variants and disease onset, correlation between genotype and phenotype is often difficult to validate (25). Moreover, the relationship between the deficiencies in the lysosomal enzyme activity and the degree of clinical severity remains unclear (26). Therefore, further evidence is needed for diagnosis of suspicious cases during new born screening.
The current standard of care for Krabbe disease patients, HSCT, only serves to delay disease progression, and is not an effective cure. However, Mikulka et al. described the combination therapies that target different pathogenic mechanisms/pathways have been more effective at reducing histological signs of disease, delaying disease onset, prolonging lifespan, and improving behavioral/cognitive functions in rodent models of Krabbe disease (27).
The newest method for measuring GALC activity is a magnetic bead based assay described by Liao et al. (26). They isolated the T lymphocytes from venous blood by magnetic bead technology. The assay used a close structural analog of the natural substrate and LC-MS/MS to quantify the amount of product. The analytical range of the assay was 20-fold greater than conventional radiometric GALC assay. Combining with genotyping, biomarker analysis, and neurological imaging, these results provide a comprehensive and fine evidence for planning for post-newborn screening follow-up for Krabbe disease.
As a recessive disease, heterozygous carriers exhibited increased risk for developing other diseases. In order to answer these question, Scott-Hewitt et al. (28) explored on cuprizone-induced myelin damage in heterozygous (GALC+/−) mice which carrying one copy of a mutation that causes Krabbe disease-like disease, and found the damage was similar in both GALC+/− and wild-type mice. However, GALC+/− mice failed to repair cuprizone-induced damage. They further found unexpected microglial defects in myelin debris clearance and Trem2 microglial protein up-regulating which was critical for debris uptake. These defects can be rescued by exposure to a lysosomal re-acidifying drug discovered, and which provides multiple clinically relevant benefits in heterozygous (GALC+/−) mouse model of Krabbe disease. Thus, heterozygous GALC mutations cause effects on biological function that may help to understand the increased disease risk in heterozygous carriers of variants and to understand why GALC variations increase the risk of MS.
Kumar et al. (29) systemic deciphered the structural and functional consequences of pathogenic missense mutations in the GALC protein causing Krabbe disease. One hundred and seventy-six mutations were retrieved from the public databases and subjected to pathogenicity, stability, and conservation analyses. Forty-five, 95, and 47 mutations to be deleterious, destabilizing, and highly conserved predicted by PredictSNP, iStable, and ConSurf. The R396L and R396W were the most deleterious and destabilizing to GALC, higher structural and functional modifications were observed on R396W in the substrate-binding site.
This case study described the case of a child with Krabbe disease and confirmed galactocerebrosidase activity deficiency. Compound heterozygous variants that caused alternative splicing or truncated proteins were identified by exome sequencing, and variants and their effects were also validated. These results provide definitive evidence for functional confirmation and clinical diagnosis. The confirmed genetic data regarding the pathogenic variants in this case could assist in genetic counseling for families and help clinicians to predict clinical outcomes and administer the clinical intervention, HSCT, for positive cases during newborn screenings in the future.
Acknowledgments
We thank the patient described in the study as well as his family members, for their cooperation.
Funding: This work was supported by the Health Commission of Henan Province (grant number: 2018020177).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-403
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tp-21-403
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-403). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the Third Affiliated Hospital of Zhengzhou University (No. 32, 2019) and informed consent was taken from parents/guardian of the patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Duffner PK, Barczykowski A, Jalal K, et al. Early infantile Krabbe disease: results of the World-Wide Krabbe Registry. Pediatr Neurol 2011;45:141-8. [Crossref] [PubMed]
- Suzuki K, Suzuki Y. Globoid cell leucodystrophy (Krabbe's disease): deficiency of galactocerebroside beta-galactosidase. Proc Natl Acad Sci U S A 1970;66:302-9. [Crossref] [PubMed]
- Beltran-Quintero ML, Bascou NA, Poe MD, et al. Early progression of Krabbe disease in patients with symptom onset between 0 and 5 months. Orphanet J Rare Dis 2019;14:46. [Crossref] [PubMed]
- Bascou N, DeRenzo A, Poe MD, et al. A prospective natural history study of Krabbe disease in a patient cohort with onset between 6 months and 3 years of life. Orphanet J Rare Dis 2018;13:126. [Crossref] [PubMed]
- Jain M, De Jesus O. Krabbe disease. In: StatPearls. Treasure Island: StatPearls Publishing, 2021.
- Bascou NA, Beltran-Quintero ML, Escolar ML. Pathogenic variants in GALC gene correlate with late onset Krabbe disease and vision loss: case series and review of literature. Front Neurol 2020;11:563724 [Crossref] [PubMed]
- Escolar ML, Poe MD, Provenzale JM, et al. Transplantation of umbilical-cord blood in babies with infantile Krabbe's disease. N Engl J Med 2005;352:2069-81. [Crossref] [PubMed]
- Wright MD, Poe MD, DeRenzo A, et al. Developmental outcomes of cord blood transplantation for Krabbe disease: a 15-year study. Neurology 2017;89:1365-72. [Crossref] [PubMed]
- Yoon IC, Bascou NA, Poe MD, et al. Long-term neurodevelopmental outcomes of hematopoietic stem cell transplantation for late-infantile Krabbe disease. Blood 2021;137:1719-30. [Crossref] [PubMed]
- Madsen AMH, Wibrand F, Lund AM, et al. Genotype and phenotype classification of 29 patients affected by Krabbe disease. JIMD Rep 2019;46:35-45. [PubMed]
- Weinstock NI, Shin D, Dhimal N, et al. Macrophages expressing GALC improve peripheral Krabbe disease by a mechanism independent of cross-correction. Neuron 2020;107:65-81.e9. [Crossref] [PubMed]
- Wiederschain G, Raghavan S, Kolodny E. Characterization of 6-hexadecanoylamino-4-methylumbelliferyl-beta-D- galactopyranoside as fluorogenic substrate of galactocerebrosidase for the diagnosis of Krabbe disease. Clin Chim Acta 1992;205:87-96. [Crossref] [PubMed]
- Raghavan S, Zeng B, Torres PA, et al. Globoid cell leukodystrophy (Krabbe disease): normal umbilical cord blood galactocerebrosidase activity and polymorphic mutations. J Inherit Metab Dis 2005;28:1005-9. [Crossref] [PubMed]
- Berardi AS, Pannuzzo G, Graziano A, et al. Pharmacological chaperones increase residual β-galactocerebrosidase activity in fibroblasts from Krabbe patients. Mol Genet Metab 2014;112:294-301. [Crossref] [PubMed]
- Giorgio A, De Stefano N. Leukoencephalopathies and metabolic diseases. Neurol Sci 2008;29:323-6. [Crossref] [PubMed]
- Abdelhalim AN, Alberico RA, Barczykowski AL, et al. Patterns of magnetic resonance imaging abnormalities in symptomatic patients with Krabbe disease correspond to phenotype. Pediatr Neurol 2014;50:127-34. [Crossref] [PubMed]
- Selleri S, Torchiana E, Pareyson D, et al. Deletion of exons 11-17 and novel mutations of the galactocerebrosidase gene in adult- and early-onset patients with Krabbe disease. J Neurol 2000;247:875-7. [Crossref] [PubMed]
- Graziano AC, Cardile V. History, genetic, and recent advances on Krabbe disease. Gene 2015;555:2-13. [Crossref] [PubMed]
- Xu C, Sakai N, Taniike M, et al. Six novel mutations detected in the GALC gene in 17 Japanese patients with Krabbe disease, and new genotype-phenotype correlation. J Hum Genet 2006;51:548-54. [Crossref] [PubMed]
- Hossain MA, Otomo T, Saito S, et al. Late-onset Krabbe disease is predominant in Japan and its mutant precursor protein undergoes more effective processing than the infantile-onset form. Gene 2014;534:144-54. [Crossref] [PubMed]
- Morais P, Adachi H, Yu YT. Suppression of nonsense mutations by new emerging technologies. Int J Mol Sci 2020;21:4394. [Crossref] [PubMed]
- Park E, Pan Z, Zhang Z, et al. The Expanding landscape of alternative splicing variation in human populations. Am J Hum Genet 2018;102:11-26. [Crossref] [PubMed]
- Cooper DN. Functional intronic polymorphisms: Buried treasure awaiting discovery within our genes. Hum Genomics 2010;4:284-8. [Crossref] [PubMed]
- Carvill GL, Engel KL, Ramamurthy A, et al. Aberrant inclusion of a poison exon causes dravet syndrome and related SCN1A-associated genetic epilepsies. Am J Hum Genet 2018;103:1022-9. [Crossref] [PubMed]
- Krieg SI, Krägeloh-Mann I, Groeschel S, et al. Natural history of Krabbe disease - a nationwide study in Germany using clinical and MRI data. Orphanet J Rare Dis 2020;15:243. [Crossref] [PubMed]
- Liao HC, Spacil Z, Ghomashchi F, et al. Lymphocyte galactocerebrosidase activity by LC-MS/MS for post-newborn screening evaluation of Krabbe disease. Clin Chem 2017;63:1363-9. [Crossref] [PubMed]
- Mikulka CR, Sands MS. Treatment for Krabbe's disease: finding the combination. J Neurosci Res 2016;94:1126-37. [Crossref] [PubMed]
- Scott-Hewitt NJ, Folts CJ, Noble MD. Heterozygous carriers of galactocerebrosidase mutations that cause Krabbe disease have impaired microglial function and defective repair of myelin damage. Neural Regen Res 2018;13:393-401. [Crossref] [PubMed]
- D TK, Jain N, Kumar S U, et al. Molecular dynamics simulations to decipher the structural and functional consequences of pathogenic missense mutations in the galactosylceramidase (GALC) protein causing Krabbe's disease. J Biomol Struct Dyn 2021;39:1795-810. [Crossref] [PubMed]
(English Language Editor: J. Gray)


