
A low abundance of genus Bacteroides in gut microbiota is negatively correlated with blood phenylalanine levels in Uygur patients with phenylketonuria
Introduction
Phenylketonuria (PKU; OMIM 261600) is a classical metabolic disorder characterized by high-plasma phenylalanine (Phe) concentration owing to pathogenic variations in the PAH gene. The first line of treatment is a life-long low-Phe diet, a lack of which results in serious neurocognitive deficits, mental health issues, and behavioral concerns (1). However, despite several decades of research, unresolved challenges remain in relation to this genetic metabolic disorder. Adherence to low- Phe diet progressively decreases after adolescence (2). Additionally, some patients remain at risk of mood disorders, anxiety, and attentional disorders (3).
Currently, few treatment options exist to reduce and maintain acceptable Phe levels. However, in addition to well-known treatable factors, such as chaperone therapy with sapropterin, Phe ammonia lyase enzyme substitution therapy, and supplementation with large neutral amino acids (LNAA), there is increasing interest in the contribution of the gut microbiota to disease, given their contribution to essential functions, such as energy harvesting, host nutrition, and immunomodulation (4). Diet-related gut microbiata alterations in PKU are well known (5,6). These two studies reported that Bacteroidetes and Firmicutes were the dominant phyla in the PKU and control groups and mild hyperphemia group, respectively. Further, they found major differences in the phylum Firmicutes. Notably, a particularly divergent response in glucose and amino acid metabolism was observed between the affected and control groups. Unlike children with PKU, Mancilla et al. (7) reported a significant differences in the abundance of specific bacterial groups in adult PKU patients. But the same was that both adults and children are involved in the abnormality of the glucose metabolism pathway.
Based on the latest available data, a total of 3 studies involving the gut microbiome with population of PKU (two children and one adults). Unlike the previous studies, our samples were Uygur children with high incidence of PKU at 1 in 5,230 children (8,9). In addition, the population consumes a high-Phe diet, including meat, pasta, and all types of nuts. It is exactly opposite with the principles of dietary treatment for PKU. Thus, this study had three aims: (I) to determine the characteristics of gut microbiota in Uygur patients with PKU; (II) to examine the correlations between these characteristics and clinical phenotypes of PKU using 16S ribosomal RNA (rRNA) sequencing; and (III) to compare the microbiota composition of PKU patients to that of healthy individuals. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-426).
Methods
Study population and sample collection
This observational study was approved by the Ethics Board of the People’s Hospital of Xinjiang Uyghur Autonomous Region (KY2021031018). All patients with PKU were recruited from the Neonatal Disease Screening Centre of the People’s Hospital of Xinjiang Uyghur Autonomous Region and written informed consent was obtained from the parents of the participants for the publication of this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A sex-, nation-, and age-matched control group of healthy children without PKU were collected from the Child Healthcare Department. To be eligible for inclusion in the PKU group, participants had to meet the following inclusion criteria: (I) have an established diagnosis of PKU; (II) accept low-Phe dietary treatment; and (III) be unrelated to other children with PKU. Individuals were excluded from both groups if they met any of the following exclusion criteria: (I) had a neurological abnormality, congenital malformation, endocrine disorder, chronic liver disease, or chronic or acute intestinal diseases; and/or (II) had been treated with antibiotics or probiotics and prebiotics (including glycomacropeptide) at any time in the 3 months preceding the study.
The following clinical parameters were evaluated: (I) sex; (II) pre-treatment Phe level; (III) current age, weight, and height; and (IV) time from birth to diagnosis. Body mass index (BMI) was calculated for all participants, and a standard BMI range according to the WHO 2009 standards was used (10). The following laboratory parameters were evaluated: the plasma levels of 11 amino acids, including Phe, tyrosine (Tyr), and free carnitine (C0). These parameters were determined using high performance liquid chromatography. The serum levels of 25-hydroxyvitamin D3 [25(OH)D3], macronutrients (carbohydrates, proteins, and lipids), amino acids, and mineral intake were calculated through a 72 h-dietary recall questionnaire. The daily intake ranges of proteins, lipids, and carbohydrates were calculated according to the Encyclopedia of Nutrition Science (11). Gut samples were collected using a stool collection kit consisting of a sterile container with solution, and a sterilized spatula (Jingyun Biotechnology Co., Ltd., Shanghai, China). The stool samples were stored at normal temperature.
The pre-treatment Phe level of the control participants ranged from 20–120 µmol/L. While those of patients with PKU on a Phe-restricted diet ranged from 120–360 µmol/L. The recommended daily Phe intake for patients with PKU in our study was 135–330 mg according to guidelines for Chinese populations (12).
DNA isolation and PCR amplification
Microbial community genomic DNA was isolated from gut samples by Kit of E.Z.N.A. Soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the instructions. The isolated DNA was electrophoresed on 1% agarose gel, using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, DE, USA) to identify the concentration and purity of DNA. Using GeneAmp 9700 polymerase chain reaction (PCR) thermal cycler (Applied Biosystems, Foster City, CA, USA) to amplify the hypervariable region (V3–V4) of the bacterial 16S rRNA gene with the primer pair 338F (5'-ACTCCTACGGGAGGCAGCAG-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3'). PCR reactions were prepared in 20 μL contained 5× TransStart FastPfu buffer (4 μL), 2.5 mM dNTPs (2 μL), 5 μM of forward primer (0.8 μL), 5 μM of reverse primer (0.8 μL), TransStart FastPfu DNA Polymerase (0.4 μL), template DNA (10 ng), and ddH2O for a final volume. Thermocycler conditions: an initial denaturation at 95 °C for 3 min, followed by 27 cycles at 95 °C for 30 s, annealing at 55 °C for 30 s, an extension at 72 °C for 45 s, and a final extension at 72 °C for 10 min. PCR reactions were performed in triplicate. PCR products were separated by electrophoresis on 2% agarose gels and purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using a Quantus Fluorometer (Promega, Madison, WI, USA).
16S rRNA gene sequencing
Purified amplicons were pooled in equimolar quantities and paired-end sequenced was performed using a MiSeq PE300 or NovaSeq PE250 (Illumina, San Diego, CA, USA) in accordance with the standard protocols by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Sequensing data analysis
We used fastp (version 0.20.0) to demultiplex and filter the raw sequencing reads (13), and merged using FLASH version 1.2.7 (14). The 300 bp reads were truncated at any site receiving an average quality score of <20 over a 50-bp sliding window, and truncated reads shorter than 50 bp and those containing ambiguous characters were discarded. Only overlapping sequences longer than 10 bp were assembled according to their overlapping sequence. These sequences were clustered into Operational Taxonomic Unit (OTU) at a 97% similarity cutoff (15) followed by UPARSE version 7.1.OTU—representing sequence was analyzed using the ribosomal database project (RDP) Classifier version 2.2 (16) against a 16S rRNA database, such as Silva v132, with a confidence threshold of 0.7.
Statistical analysis
Comparisons of linear relationship between phenotypes and gut microbiota composition was performed using the lm function in R’s regression analysis package. For evaluating Spearman-rank correlation analysis between phenotype and microbiota pathways, the cor and cor.test functions in the R package was performed. False discovery rate (FDR)-corrected P values <0.05 and Spearman correlation coefficient absolute values >0.3 were considered as significant.
Results
Participant characteristics
Of the 11 patients who completed the study, 7 were female. Patients’ ages ranged from 10 months to 8 years and 3 months, and patients had a BMI of 16.36±0.35 kg/m2, and a mean pre-treatment plasma Phe level of 986.01±168.19 µmol/L. All patients were on a low-Phe- diet supplemented with a metabolic formula (protein substitute) without prebiotics, and no one received tetrahydrobiopterin (BH4), LNAA-, or glycomacropeptide-based treatment. The mean plasma Phe level on the day on which the blood samples were drawn was 481.46±121.87 µmol/L, and the mean daily intake of Phe was 577.72±127.09 mg/d. Table 1 summarizes the values for other blood indicators and nutrients from daily intake (see available online: https://cdn.amegroups.cn/static/public/tp-21-426-1.xlsx).
Table 1
| No. | Sex | Age (months) | BMI (kg/m2) | Age at diagnosis (days) | Pre-treatment Phe (µmol/L) | Blood sample | Daily intake | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 25(OH)D3 | Phe (µmol/L) | Phe (mg) | Protein (g) | Carbohydrate (g) | |||||||
| Standard | – | – | 15.8–17 | – | 20–120 | >30 | 120–360 | 135–330 | 20–40 | 60–120 | |
| 1 | Female | 75 | 15.42 | 93 | 776.4 | 21.89 | 240.29 | 864.37 | 42.47 | 215.3 | |
| 2 | Female | 14 | 17.91 | 90 | 418.2 | 29.48 | 237.65 | 905.33 | 54.67 | 276.87 | |
| 3 | Female | 78 | 15.47 | 288 | 865.8 | 24.56 | 230.33 | 729.33 | 35.53 | 190.47 | |
| 4 | Female | 51 | 16.49 | 64 | 1,694.4 | 33.68 | 691.99 | 127 | 26.17 | 191 | |
| 5 | Male | 10 | 17.53 | 57 | 888.6 | 60.52 | 272.46 | 89.67 | 16.2 | 73.43 | |
| 6 | Male | 35 | 16.33 | 330 | 1,680 | 18.32 | 1,581.9 | 232.67 | 29.87 | 179.37 | |
| 7 | Female | 100 | 17.36 | 890 | 1,655.4 | 17.48 | 480.88 | 200.67 | 28.6 | 162.4 | |
| 8 | Male | 67 | 15 | 138 | 1,582.8 | 23.08 | 654.87 | 102.5 | 20.73 | 144.93 | |
| 9 | Male | 19 | 16.77 | 141 | 432.6 | 20.03 | 168.36 | 963.67 | 36.9 | 232.27 | |
| 10 | Female | 43 | 14.35 | 284 | 416.4 | 33.65 | 351.67 | 1,033.67 | 35.1 | 207.13 | |
| 11 | Female | 47 | 17.35 | 122 | 435.6 | 25.5 | 385.69 | 1,106 | 41.8 | 242.83 | |
| Mean ± SEM | – | 49±8.70 | 16.36±0.35 | 227±72.34 | 986.01±168.19 | 28.02±3.65 | 481.46±121.87 | 577.72±127.09 | 33.46±3.26 | 192.36±16.37 | |
PKU, phenylketonuria; BMI, body mass index; Phe, phenylalanine; 25(OH)D3, 25-hydroxyvitamin D3; SEM, standard error of the mean.
Comparison of ecological diversity of gut microbiota
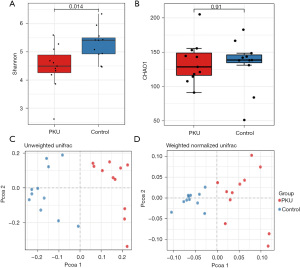
After filtering reads by removing sequencing artifacts, a total of 722,283 high-quality sequences with an average of 16,186 sequences per sample were retained. The alpha-diversity analysis revealed no significant differences between the PKU and control groups based on the Ace (P=0.95) and Chao1 (P=0.8) indices; however, significant differences were found with the Simpson (P=0.04) and Shannon (P=0.014) indices, which implies that the control group had a higher diversity of gut microbiota than the PKU group (see Figure 1A,1B). Additionally, the beta-diversity analysis showed that the structure of the fecal microbiota of the PKU group differed to that of the control group according to both unweighted and weighted UniFrac distances (see Figure 1C,1D).
Comparison of gut microbiota composition
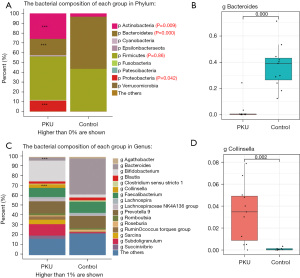
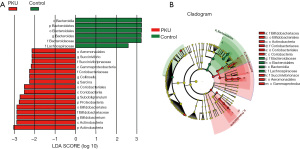
At the phylum level, we detected more than 13 known types of bacteria, the most populous being Firmicutes (44.3%), Actinobacteria (25.9%), Bacteroidetes (16.6%), and Proteobacteria (10.9%) in the PKU group, and Bacteroidetes (53.2%), Firmicutes (43.1%), Actinobacteria (3.3%), and Proteobacteria (0.12%) in the control group. The bacterial abundance between the two groups differed significantly (PActinobacteria=0.009, PBacteroidetes<0.001, and PProteobacteria=0.042), except for that of Firmicutes (PFirmicutes=0.86) (see Figure 2A). Next, we chose the most populous bacteria of each group for the genus-level analysis. In the PKU group, the most populous genera were Bifidobacterium (19.7%), Prevotella (11.5%), Subdoligranulum (10.7%), and Faecalibacterium (8.4%), whereas Bacteroides (36.6%), Prevotella (12.5%), Faecalibacterium (11.3%), and Bifidobacterium (3.2%) were the most populous in the control group (see Figure 2B). Further, we found that the abundance of Bacteroides (2.7% vs. 36.6%; P<0.001) and Collinsella (3.38% vs. 0.09%; P=0.002) differed significantly between the PKU and control groups (see Figure 2C,2D). The composition of gut microbiota for each child at the phylum and genus level is presented in Figure S1, and the differences in gut microbiota between the two groups are represented graphically in Figure 3.
Microbial function prediction between the groups
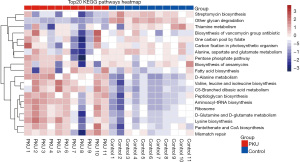
On the basis of the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis, we observed at least 20 differentially abundant bacterial functions related to metabolism between the PKU and control groups (all P<0.05). We found that all samples of the PKU group were significantly decreased in the streptomycin biosynthesis and glycan degradation. In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis predicted a decrease in glutamate metabolism in the PKU group (see Figure 4).
Associations between clinical phenotype and microbiota at the genus level
The correlations between the clinical phenotype and gut bacteria of our PKU cohort are summarized in Table 2 and available online: https://cdn.amegroups.cn/static/public/tp-21-426-2.xlsx. We found a strong negative correlation between the genus Bacteroides and blood Phe level (P=0.014); this bacterium was the least populous in the PKU group. The genera Agathobacter and Fusicatenibacter had low abundance and were associated with neurological abnormalities in the PKU group (P=0.005 and P=0.006, respectively). We also found that the genus Prevotella was associated with the daily intake of various nutrients, specifically, Phe, calories, protein, fat, and fiber.
Table 2
| Category | Actinobacteria | Bacteroidetes | Firmicutes | Proteobacteria | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bifidobacterium | Collinsella | Bacteroides | Prevotella | Agathobacter | Fusicatenibacter | Roseburia | Terrisporobacter | ||||
| Clinical information | |||||||||||
| Neurological abnormality | 0.56 | 0.85 | 0.333 | 0.704 | 0.005* | 0.006* | 0.583 | 0.322 | 0.257 | ||
| Blood sample | |||||||||||
| 25(OH)D3 | 0.189 | 0.832 | 0.611 | 0.852 | 0.353 | 0.852 | 0.016* | 0.366 | 0.361 | ||
| Plasma Phe | 0.79 | 0.957 | 0.014* | 0.649 | 0.31 | 0.325 | 0.749 | 0.574 | 0.295 | ||
| Plasma Tyr | 0.65 | 0.083 | 0.852 | 0.183 | 0.527 | 0.957 | 0.2 | 0.935 | 0.361 | ||
| Plasma C0 | 0.417 | 0.055 | 0.16 | 0.094 | 0.957 | 0.169 | 0.142 | 0.967 | 0.877 | ||
| Daily intake | |||||||||||
| Phe | 0.484 | 0.831 | 0.79 | 0.008* | 0.718 | 0.729 | 0.611 | 0.093 | 0.162 | ||
| Calories | 0.709 | 0.573 | 0.519 | 0.015* | 0.62 | 0.831 | 0.709 | 0.007* | 0.112 | ||
| Protein | 0.769 | 0.611 | 0.385 | 0.029* | 0.678 | 0.873 | 0.519 | 0.008* | 0.121 | ||
| Fat | 0.65 | 0.811 | 0.117 | 0.025* | 0.852 | 0.79 | 0.915 | 0.08 | 0.011* | ||
| Carbohydrate | 0.555 | 0.689 | 0.831 | 0.062 | 0.474 | 0.611 | 0.873 | 0.038* | 0.279 | ||
| Fiber | 0.915 | 0.631 | 0.894 | 0.036* | 0.545 | 0.669 | 0.811 | 0.107 | 0.104 | ||
*, statistically significant (P<0.05). PKU, phenylketonuria; 25(OH)D3, 25-hydroxyvitamin D3; Phe, phenylalanine; Tyr, tyrosine; C0, free carnitine.
Discussion
Our patients were Uyghurs, who had distinct culture characteristics driven by the ancient Silk Road, an important trade route that connected culture and trade among China, Europe, and Asia (8). This ethnic population has a high incidence of PKU and has distinctly different eating habits to those of the Han population in China. Thus, we sought to determine characteristics of the gut flora of individuals with PKU from this population.
In this study, we used a safe, non-invasive method to collect stool samples from patients with PKU and healthy control individuals, and found that the diversity of microbiota in patients with PKU was significantly decreased compared to that of control participants. Ecological theories predict that a decreased diversity of gut microbial significantly limits the ability of the microbiome to withstand major shifts, potentially leading to alternate, diseased, or stable states (17,18). In contrast to the control group and findings of previous studies (5,6), in the present study, Firmicutes (44.3%) and Actinobacteria (25.9%) were the most populous phyla in the PKU group. In the healthy control group, Firmicutes and Bacteroidetes accounted for more than 90% of the gut microbiome, followed by minor abundances of Actinobacteria and Proteobacteria. We speculate that this is a common effect of dietary habits and disease itself. There may be several reasons for these differences.
First, in our study, the genus Collinsella, which belongs to the phyla Actinobacteria, was significantly more populous in the PKU group. Studies have shown that low dietary fiber intake increases the abundance of Collinsella in the gut microbiome (19-21). This finding is consistent with the eating pattern of the Uygur population, who prefer to consume meat and wheat-based foods (8). Second, we followed-up these patients of PKU who have high abundance of genus Bifidobacterium. We found six of them still had breast milk sometimes even they were more than 2 years. This consistent with the features of longer breastfeeding time of Uyghur mothers (22). In addition, we found that the genus Bacteroides were significantly lower in our PKU cohort. This was the main reason for the reduced abundance of phyla Bacteroidetes, and lower abundance of Bacteroidetes was negatively correlated with blood Phe level in our affected cohort. There is increasing evidence that Bacteroides have a remarkable capability to use complex recalcitrant glycans, and thus may have the capability to use diverse carbon sources by acquiring enzymes via heterologous gene transfer events in the gut environment (23,24). This reasonably explains why the ability of glycan degradation in the PKU group was decreased as predicted in the KEGG pathway analysis (see Figure 4). Additionally, this result is also in line with previous reports that the daily glycemic index is higher in children with PKU (5,6). However, the increased glucose in these studies may be caused by phyla Firmicutes, whereas ours may be caused by phyla Bacteroidetes.
Theoretically, genus Prevotella (belong to phylum Bacteroidetes) should be highly abundant in children who consumed diets with high amounts of plant polysaccharides and fiber, lower in animal protein and saturated fats (25,26), which similar with the dietary pattern of children with PKU. Considering that children with PKU are in the growth and development period, adequate protein intake must be ensured. The patients need to restrict the intake of natural protein, an important source of protein is the amino acid formula of free-Phe. After absorbing protein in small intestine, specific carbohydrates which was lacking oligosaccharides, lactose polymers into the large intestine (11). The carbohydrates reduce the abundance of Lactic acid bacteria, Clostridium butyricum and Prausnitzii (27), which affect the synthesis of short-chain fatty acids (6). We need to analyze the changes of intestinal flora in combination with many environmental factors.
Based on microbial function prediction using the PICRUSt analysis, we focused on glutamate metabolism, as we predicted that it would be decreased in the PKU group. Glutamate metabolism involves enterocyte proliferation, suppresses proinflammatory signaling pathways, confers protection against apoptosis and cellular stresses during normal and pathological conditions, including autism and other conditions or diseases related to the central nervous system (28,29). During the follow-up of our PKU cohort, we found that 27% of the children exhibited cognitive impairment and developmental delay even under strict Phe-restricted diets. Further, our data were consistent with those of a systematic review (25–33%) (30). In relation to the role of glutamate in the gut, future studies of neurotransmitter metabolomics need to be conducted to confirm the status of glutamate metabolism in the intestine of children with PKU.
Limitations
We found evidence of a strong negative correlation between the genus Bacteroides and blood Phe levels in our PKU group, which corresponds to the low abundance of this bacteria described earlier. As bacterial abundance is readily affected by environmental factors, we were unable to determine whether the low abundance of this bacteria is a cause or a consequence of blood Phe level in our cohort. In addition, this study comprised patients with a specific ethnic and genetic background. Future studies should increase the sample size and add different races and a mild hyperphemia group as control groups to verify the level of genus Bacteroides in different groups. In addition, we can better explain the relationship between gut microbiome and diet habits, genetic background, diet treatment and the disease itself.
Conclusions
Through high-throughput sequencing of the 16S rRNA gene, we discovered that the genus Bacteroides were not only in extremely low abundance in Uygur patients with PKU, but were also negatively correlated with blood Phe level. Our findings predicted a decreased ability of glutamate metabolism in the PKU group. Additionally, in PKU, because the treatment interventions are based on long-term or lifelong customized dietary plans. According to the only study on adult with PKU, the impact of the PKU diet and treatment over the course of a patient’s lifetime remains an unknown. Interestingly, these studies (include ours) all indicated significant differences between the PKU and the control group in glucose metabolism or synthesis, which could be the direction of follow-up studies. In future studies, and as part of efforts to increase the sample size, we will refine the blood and diet indicators related to glucose metabolism to better understand the underlying causes of and confirm the reasons for abnormal glucose metabolism. We will also perform microbiota metabolomic research to investigate glutamate levels in the gut microbiota of children with PKU who have developmental delays.
Acknowledgments
We are very grateful to the patients and their families, the clinicians who took care of the patients, and our team members who contributed to this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-426
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tp-21-426
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-426). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of the People’s Hospital of Xinjiang Uyghur Autonomous Region (KY2021031018), and written informed consent was obtained from the parents of the participants for the publication of this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lowe TB, DeLuca J, Arnold GL. Similarities and differences in key diagnosis, treatment, and management approaches for PAH deficiency in the United States and Europe. Orphanet J Rare Dis 2020;15:266. [Crossref] [PubMed]
- Cazzorla C, Bensi G, Biasucci G, et al. Living with phenylketonuria in adulthood: the PKU ATTITUDE study. Mol Genet Metab Rep 2018;16:39-45. [Crossref] [PubMed]
- Bilder DA, Noel JK, Baker ER, et al. Systematic review and meta-analysis of neuropsychiatric symptoms and executive functioning in adults with phenylketonuria. Dev Neuropsychol 2016;41:245-60. [Crossref] [PubMed]
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369-79. [Crossref] [PubMed]
- Pinheiro de Oliveira F, Mendes RH, Dobbler PT, et al. Phenylketonuria and gut microbiota: a controlled study based on next-generation sequencing. PLoS One 2016;11:e0157513 [Crossref] [PubMed]
- Bassanini G, Ceccarani C, Borgo F, et al. Phenylketonuria diet promotes shifts in firmicutes populations. Front Cell Infect Microbiol 2019;9:101. [Crossref] [PubMed]
- Mancilla VJ, Mann AE, Zhang Y, et al. The adult phenylketonuria (PKU) gut microbiome. Microorganisms 2021;9:530. [Crossref] [PubMed]
- Su Y, Wang H, Rejiafu N, et al. The molecular epidemiology of hyperphenylalaninemia in Uygur population: incidence from newborn screening and mutational spectra. Ann Transl Med 2019;7:258. [Crossref] [PubMed]
- Hillert A, Anikster Y, Belanger-Quintana A, et al. The genetic landscape and epidemiology of phenylketonuria. Am J Hum Genet 2020;107:234-50. [Crossref] [PubMed]
- World Health Organization. WHO child growth standards and the identification of severe acute malnutrition in infants and children: a joint statement by the World Health Organization and the United Nations Children’s Fund. 2009. Available online: https://apps.who.int/iris/bitstream/handle/10665/44129/9789241598163_eng.pdf?ua=1
- Yang YX Ge YK. Encyclopedia of nutrition science. 2nd ed. Beijing: People’s Medical Publishing House, 2019.
- Subspecialty Group of Newborn Screening, Society of Birth Defects Prevention and Control, Chinese Preventive Medicine Association. Consensus statement on dietary treatment and nutritional management for phenylalanine hydroxylase deficiency. Zhonghua Er Ke Za Zhi 2019;57:405-9. [PubMed]
- Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884-90. [Crossref] [PubMed]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957-63. [Crossref] [PubMed]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. [Crossref] [PubMed]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261-7. [Crossref] [PubMed]
- Mallon CA, Le Roux X, van Doorn GS, et al. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader's niche. ISME J 2018;12:728-41. [Crossref] [PubMed]
- Mallon CA, Poly F, Le Roux X, et al. Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology 2015;96:915-26. [Crossref] [PubMed]
- Frost F, Storck LJ, Kacprowski T, et al. A structured weight loss program increases gut microbiota phylogenetic diversity and reduces levels of Collinsella in obese type 2 diabetics: a pilot study. PLoS One 2019;14:e0219489 [Crossref] [PubMed]
- Gomez-Arango LF, Barrett HL, Wilkinson SA, et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 2018;9:189-201. [Crossref] [PubMed]
- Karlsson FH, Fåk F, Nookaew I, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012;3:1245. [Crossref] [PubMed]
- Cheng F. Epidemiology Study of Risk Factors and Genetic Susceptibility of Han and Uigur Breast Cancer in Xinjiang. Urumqi: Xinjiang Medical University, 2010.
- Luis AS, Briggs J, Zhang X, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 2018;3:210-9. [Crossref] [PubMed]
- Ndeh D, Rogowski A, Cartmell A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017;544:65-70. [Crossref] [PubMed]
- Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066-71. [Crossref] [PubMed]
- Miura K, Ohnishi H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J Gastroenterol 2014;20:7381-91. [Crossref] [PubMed]
- Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 2019;7:91. [Crossref] [PubMed]
- Perna S, Alalwan TA, Alaali Z, et al. The role of glutamine in the complex interaction between gut microbiota and health: a narrative review. Int J Mol Sci 2019;20:5232. [Crossref] [PubMed]
- Wang M, Wan J, Rong H, et al. Alterations in gut glutamate metabolism associated with changes in gut microbiota composition in children with autism spectrum disorder. mSystems 2019;4:e00321-18. [Crossref] [PubMed]
- Burlina AP, Lachmann RH, Manara R, et al. The neurological and psychological phenotype of adult patients with early-treated phenylketonuria: a systematic review. J Inherit Metab Dis 2019;42:209-19. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


