
A novel de novo TBX20 variant in a 6-year-old Chinese girl with left ventricular noncompaction: a case report
Introduction
Left ventricular noncompaction (LVNC) is a particular type of cardiomyopathy that develops due to absent or incomplete myocardial compaction during embryonic morphogenesis, causing excessively prominent trabecular meshwork and deep intertrabecular recesses to develop in the left ventricle (LV). The incidence rate for LVNC was reported to be between 0.05/100/year and 0.25/100/year (1) and it is estimated that at least half the cases of LVNCs are hereditary (2). In 2006, the American Heart Association (AHA) classified LVNC as a primary (hereditary) cardiomyopathy (3). LVNC occurs at all ages and the clinical manifestations are highly variable, ranging from no symptom to congestive heart failure, arrhythmia, thrombosis, and even sudden cardiac death. In this case, we identified a novel de novo TBX20 variant (c.859C>T) in a Chinese child diagnosed with LVNC using exome sequencing (ES).
We present the following case in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-21-460/rc).
Case presentation
A 6-year-old Chinese girl was admitted to our hospital for “loss of consciousness”. The child reportedly felt fatigued while walking and then fainted. Her symptoms included unresponsiveness, tetraplegia, eyes-closing, and trauma from her head hitting the ground. The patient then developed rigidity in her limbs, which lasted for about 1 minute and resolved spontaneously. She was in good spirits after the episode.
Following this, the girl visited our outpatient clinic, and an unenhanced cranial computed tomography scan showed a low-density area in the right temporal lobe. She was admitted to the hospital for “syncope of unknown origin for further diagnosis and treatment”.
The girl was in a good state of physical fitness. In the month before admission, she reported several episodes of dizziness without blurred vision, which lasted less than one minute and resolved spontaneously.
The girl (G1P1) was born via cesarean delivery at a full-term gestational age with a normal birth history. She had good academic performance and presented normal growth and development. There was no known family history of syncope, congenital heart disorders, primary cardiomyopathies, sudden death, miscarriage and early childhood death etc.
Her general medical examination was unremarkable. On initial physical examination, she had a blood pressure of 90/60 mmHg with a pulse of 100 beats per minute (bpm) and respiration of 20 bpm. Her height (121 cm) and weight (18.5 kg) were normal for a girl of her age. She was energetic and conscious; her face was ruddy and healthy-looking. She had clear lungs and normal heart sounds with no murmur or gallop on auscultation. Her lower extremities were warm to touch without edema. The liver and spleen were not palpable or enlarged. Her nervous system examination showed no obvious positive signs.
All procedures performed in studies involving human participants were conducted in accordance with the ethical standards of the institutional and/or national research committee(s) and the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s parents for publication of this case report. A copy of the written consent is available for review by the editorial office of this journal.
Laboratory examinations
The blood routine test, serum myocardial enzymes spectrum, blood gas analysis, level of blood lactate, the coagulation function test, and the liver and kidney function test were within normal limits.
Imaging examinations
2D-echocardiography revealed a dilated LV (LV diastolic dimension of 45 mm, LV systolic dimension of 32 mm, Z score was 3.8) with numerous prominent trabeculations, and a two-layered structure, comprising a compacted thin epicardial band (3.6 mm) and a much thicker non-compacted endocardial layer (11 mm) of trabecular meshwork, with deep endomyocardial spaces and deep intertrabecular recesses in the lower and middle LV. The left atrium (LA) was obviously enlarged, the right atrium (RA) and right ventricle (RV) were slightly enlarged. The LV posterior wall motion was reduced, and the LV ejection fraction (EF) was 57%. Doppler detected moderate mitral valve regurgitation (P=48 mmHg) and mild tricuspid valve regurgitation (P=38 mmHg) (Figure 1).
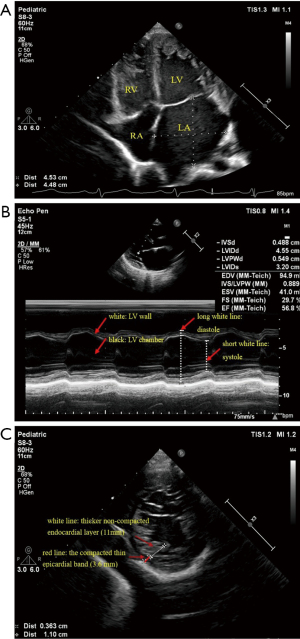
The cardiac magnetic resonance (CMR) examination showed a dilated LV and LA in systole, some prominent trabeculations, and deep perfused intertrabecular recesses, which were seen in the LV apex and lateral wall with a compacted thin epicardial band (2.9 mm) and a thicker non-compacted endocardial layer (8.5 mm). Cine showed blood communication between intertrabecular recesses and the heart cavity (Figure 2).

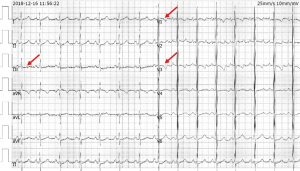
The standard 12-lead electrocardiograph (ECG) showed T-wave morphology changes in several leads (Figure 3). Arrhythmia was not recorded on 24-h Holter monitoring.
Brain magnetic resonance imaging (MRI), brain magnetic resonance angiography (MRA), brain magnetic resonance venography (MRV), electroencephalogram (EEG), and 16-h video EEG showed no apparent abnormality.
Molecular findings
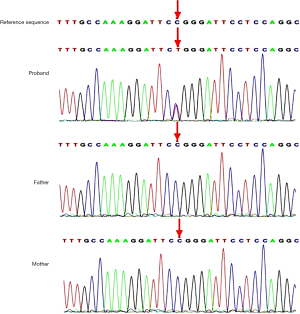
The karyotype of the proband was normal. After obtaining the patient’s consent, we performed trio-based ES on the patient and her healthy parents. Genomic DNA was extracted from peripheral blood. The sequence variants were functionally annotated and filtered using known populations and databases, including 1,000 genomes, Single Nucleotide Polymorphism Database, Genome Aggregation Database, Clin Var, Human Gene Mutation Database, and Online Mendelian Inheritance in Man. Candidate variants were then evaluated in the context of clinical presentation and inheritance mode. Unreported nonsynonymous single nucleotide variants were predicted using in silico predictive algorithms such as Mutation Taster (http://www.mutationtaster.org), Sorting Intolerant from Tolerant (http://sift.jcvi.org), Polymorphism Phenotyping v2 (http://genetics.bwh.harvard.edu/pph2/), and phyloP (http://compgen.bscb.cornell.edu/phast/) to evaluate any potentially damaging effects to the protein function. Putative pathogenic variants were subsequently validated by Sanger sequencing. Finally, we filtered out variants in genes unrelated to “cardiac dilatation”. The genetic analysis in the proband identified a de novo heterozygous variant in exon 9 of the TBX20 gene (NM_001077653, c.859C>T), this missense mutation caused the substitution (Arg→Trp) at position 287 of the protein sequence. Sanger sequencing failed to detect this variant in the parents (Figure 4). The identified variant was so low-frequency in the population that it was even absent from EVS, ExAC, gnomAD and dbSNP databases. It was predicted as deleterious by five algorithms (PROVEAN score 7.34, SIFT score 0.009, REVEL score 0.882, M-CAP score 0.558, CADD score 32). Two algorithms predicted it as probably damaging (Polyphen_HDIV score 1, Polyphen_HVAR score 1), and it was predicted as “disease causing” by mutationtaster with a score of 1. According to the 2015 ACMG guidelines (4), the variant was classified as a likely pathogenic variant (PS2 + PM1 + PM2 + PP3).
Diagnosis and treatment
The patient was diagnosed with LVNC and suspected cardiac syncope. She received oral therapy including digoxin (9 µg/kg/d, bid), spironolactone (1 mg/kg/d, bid), captopril (1 mg/kg/d, tid), coenzyme Q, and vitamin C.
Outcome and follow-up
The patient has been followed up approximately every 3 months for regular cardiology. She has stable symptoms with New York Heart Association functional class I and no complaint, including syncope. After 2 years of follow-up, the last 2D-echocardiography still showed dilated LV (LV diastolic dimension of 46.4 mm, LV diastolic systolic dimension of 31.7 mm, Z score of 3.5) and LA. The non-compacted endocardial layer was thicker (16.6 mm) and the compacted thin epicardial was thinner (3 mm) than before. The basal segments of the interventricular septum thinned, the LV posterior wall motion was slightly reduced, and the LVEF increased to 60%. She has not shown any obvious clinical signs or symptoms.
Discussion
There is currently no diagnostic gold standard for LVNC. Echocardiography is the first choice and the most commonly used cardiac imaging method for diagnosis, the most widely used diagnostic criteria of echocardiography were proposed by Jenni in 2001 (5). According to Jenni criteria, the diagnosis of LVNC was established in this case.
The first symptoms of LVNC onset in every age group, and the clinical presentation is non-specific and various. Some patients are lifelong asymptomatic. Syncope without arrhythmia as first manifestation of LVNC was quite rare in previous cases. In this case, we could not rule out the possibility of neutrally mediated syncope and unexplained syncope judging from the ECG, EEG, and brain MRI scanning results as there were no obvious abnormalities. According to the typical presentation of LVNC on the echocardiography, CMR, and the low-normal EF in this patient, cardiac syncope could not be entirely excluded. Because of the child’s young age, some other manifestations (e.g., heart failure, thrombosis, arrhythmia) possibly occurred during follow-up.
With the development of next-generation sequencing technologies, our knowledge about regulating gene expression in LVNC is growing. More and more variants have been shown to cause LVNC, such as on sarcomere protein genes (e.g., MYH7, ACTCl, TPMl, TNNT2, MYBPC3), ion channel genes (e.g., ANK2, KCNH2, KCNE3) and transcription factor gene (e.g., PRDM16). All those variants have not identified in this case. However, unknown variants account for about 20% of LVNC families (6). The presence of pathogenic variants is an independent risk factor for the poor prognosis in patients with LVNC (7).
The TBX20 gene is localized to human chromosome 7p14.3 and contains 8 exons. It encodes a transcription factor protein consisting of 447 amino acid residues. The amino acids between positions 289 to 888 compose the T-box DNA-binding domains, which bind to specific DNA sequences and then lead to transcriptional regulation. At least seven members of the T-box gene family are expressed in human embryonic and vertebrate hearts, such as TBX1 to TBX5, TBX18, and TBX20 (8). The TBX20 gene has strong transcriptional activation and repression domains, which interact with other transcription factors such as NKX25, GATA4, and TBX5 that regulate the development of the embryonic heart (9).
It has been reported that TBX20 was related to congenital heart disease, heart valve development and dilated cardiomyopathy (DCM) (9,10). Animal experimental studies have also confirmed that TBX20 allele variants lead to DCM (11,12). In 2017, American researchers discovered that LVNC carried the TBX5 variant for the first time (13). So far, studies relating to LVNC carrying TBX20 gene variant are still rare. In 2016, Kodo et al. reported an LVNC family with TBX20 nonsense variant (c.951C>A, p.Y317*) and a case of LVNC patient with TBX20 (c.785C>T, p.T262M) de novo variant; this study also found that cardiomyocytes differentiated from human induced pluripotent stem cells in LVNC patients with TBX20 variants that had abnormal TGF-β signaling pathways; TGF-β signaling pathways are necessary for the development of the heart’s compacted endocardial layer (14). In 2017, American researchers performed gene sequencing in 190 LVNC patients and found that a gene variant in TBX20 was present in one patient, but the specific variant site was not reported (15).
This case presented a pediatric LVNC patient with TBX20 [c.859C>T, p.(Arg287Trp)] gene variant, which caused the substitution at amino acid 287[p.(Arg287Trp)]. The parents did not show this variant, and thus, it was considered a de novo variant, we suggest that the LVNC in this patient is resulted from the TBX20 variant.
In conclusion, the de novo variant of TBX20 in LVNC expands the spectrum of variants that cause LVNC and contributes to the genetic counseling and individualized treatment of patients. Clinicians should focus on exploring the clinical and genetic characteristics of LVNC to provide therapies and follow-up to improve the outcome. Further validation of the function of the gene is also required in future studies.
Acknowledgments
We are thankful to the members of the family for their participation and help in the study.
Funding: This project was supported by the Project of Science and Technology for People’s Livelihood of Suzhou of Science (No. SS201871) and Science and Education Strong Health Project in Suzhou (No. KJXW2020023).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-21-460/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-21-460/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s parents for publication of this case report. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Arbustini E, Weidemann F, Hall JL. Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol 2014;64:1840-50. [Crossref] [PubMed]
- Sedaghat-Hamedani F, Haas J, Zhu F, et al. Clinical genetics and outcome of left ventricular non-compaction cardiomyopathy. Eur Heart J 2017;38:3449-60. [Crossref] [PubMed]
- Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation 2006;113:1807-16. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Jenni R, Oechslin E, Schneider J, et al. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart 2001;86:666-71. [Crossref] [PubMed]
- Monserrat L, Hermida-Prieto M, Fernandez X, et al. Mutation in the alpha-cardiac actin gene associated with apical hypertrophic cardiomyopathy, left ventricular non-compaction, and septal defects. Eur Heart J 2007;28:1953-61. [Crossref] [PubMed]
- Li S, Zhang C, Liu N, et al. Genotype-positive status is associated with poor prognoses in patients with left ventricular noncompaction cardiomyopathy. J Am Heart Assoc 2018;7:e009910. [Crossref] [PubMed]
- Steimle JD, Moskowitz IP. TBX5: a key regulator of heart development. Curr Top Dev Biol 2017;122:195-221. [Crossref] [PubMed]
- Liu P, Sun Y, Qiu G, et al. Silencing of TBX20 gene expression in rat myocardial and human embryonic kidney cells leads to cell cycle arrest in G2 phase. Mol Med Rep 2016;14:2904-14. [Crossref] [PubMed]
- Qian L, Mohapatra B, Akasaka T, et al. Transcription factor neuromancer/TBX20 is required for cardiac function in drosophila with implications for human heart disease. Proc Natl Acad Sci U S A 2008;105:19833-8. [Crossref] [PubMed]
- Zhao CM. TBX20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med 2016;54:325-32. [Crossref] [PubMed]
- Lu F, Langenbacher A, Chen JN. Tbx20 drives cardiac progenitor formation and cardiomyocyte proliferation in zebrafish. Dev Biol 2017;421:139-48. [Crossref] [PubMed]
- Wang C, Hata Y, Hirono K, et al. A wide and specific spectrum of genetic variants and genotype-phenotype correlations revealed by next-generation sequencing in patients with left ventricular noncompaction. J Am Heart Assoc 2017;6:006210. [Crossref] [PubMed]
- Kodo K, Ong SG, Jahanbani F, et al. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol 2016;18:1031-42. [Crossref] [PubMed]
- Miszalski-Jamka K, Jefferies JL, Mazur W, et al. Novel genetic triggers and genotype-phenotype correlations in patients with left ventricular noncompaction. Circ Cardiovasc Genet 2017;10:e001763. [Crossref] [PubMed]


