
Study of novel NARS2 variants in patient of combined oxidative phosphorylation deficiency 24
Introduction
NARS2, the gene encoding mitochondrial asparaginyl-tRNA synthetase 2, is located on 11q14.1 and comprises 14 exons spread over 139 kb of DNA (1). NARS2 is a member of the mitochondrial aminoacyl-tRNA synthetases (mt-aaRSs), a key component of the mitochondrial translation apparatus. mt-aaRSs are encoded by nuclear DNA and then transported to the mitochondria (2). Each mt-aaRS binds a specific mitochondrial tRNA with its cognate amino acid (3). For example, NARS2 catalyzes the attachment of asparagine amino acids to mitochondrial tRNAAsn. This aminoacylation is essential for efficient mitochondrial protein synthesis, including four subunits of the mitochondrial oxidative phosphorylation (OXPHOS) complex (4).
Biallelic variants in NARS2 are associated with combined oxidative phosphorylation deficiency 24 (COXPD24; MIM: 616239) and autosomal recessive deafness-94 (MIM: 618434). COXPD24 is an autosomal recessive disorder characterized by early-onset seizures, global developmental delay, impaired intellectual development, myopathy, hypotonia, and hearing impairment. Some patients also show abnormalities in the kidney and/or liver (5-7). Autosomal recessive deafness-94 causes patients to exhibit bilateral sensorineural hearing loss (8). Despite the wide spectrum of disease phenotypes stemming from NARS2 variants, the genotype-phenotype relationships remain unpredictable due to a limited number of known cases. Furthermore, the underlying mechanisms of NARS2-related disease have not been comprehensively studied.
Here, we present the clinical and genetic features of a patient with compound heterozygous variants in NARS2. We used both in silico and in vitro experiments to investigate the functional impact of NARS2 variants on the NARS2 protein. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-21-570/rc).
Methods
Patient
The patient was a 6-month-old Chinese boy, the first child to a healthy, non-consanguineous couple. No relevant family history was present. The patient’s prenatal history was unremarkable. He was full-term at birth, with normal weight and length. At 3 months old, the patient developed generalized epilepsy and myoclonic seizures. He could not lift his head up stably, indicating early developmental delay. Muscle weakness and hypotonia were noted during the physical examination.
Electroencephalography (EEG) showed arrhythmic slow waves mixed with irregular spikes, as well as sharp slow waves in the central, parietal, and temporal regions. Magnetic resonance imaging of the brain was normal at 3 months. An auditory brainstem response test revealed severe bilateral hearing impairment. Laboratory tests showed increased serum levels of alanine transaminase [ALT: 67 U/L (N, 5–40 U/L)], aspartate transaminase [AST: 77 U/L (N, 5–40 U/L)], gamma-glutamyltranspeptidase [GGT: 148 U/L (N, 7–50 U/L)], alkaline phosphatase [ALP: 436 U/L (N, 50–400 U/L)], α-hydroxybutyrate dehydrogenase [α-HBDH: 477 U/L (N, 95–250 U/L)], creatine kinase-MB [CK-MB: 62 U/L (N, 0–24 U/L)], lactate dehydrogenase [LDH: 569 U/L (N, 80–285 U/L)], and troponin I [TnI: 0.06 ng/mL (N, 0–0.03 ng/mL)]. Serum electrolyte levels, lipid profile, and biochemical characteristics of the kidney were within the normal range. Metabolic screening of blood and urine revealed no abnormalities. Blood and cerebrospinal fluid (CSF) cultures were negative. Laboratory tests excluded pathogenic microorganisms, such as Chlamydia pneumoniae, Mycoplasma pneumoniae, adenovirus, respiratory syncytial virus, influenza virus, and Epstein-Barr virus. Muscle biopsy study was not performed, because it is an invasive test and the parents refused. The patient was treated with anti-seizure medications, including phenobarbital, midazolam, topiramate, clonazepam, and sodium valproate. These drugs reduced epilepsy frequency and amplitude, but partial onset seizures still occurred. The patient died of a seizure at 6 months of age.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of SCMC (No. SCMCIRB-K2020060-1) and informed consent was taken from the patient’s parents prior to the study.
Genetic investigation
Genomic DNA was extracted from peripheral blood leukocytes of the patient and his parents. Whole exome sequencing (WES) was performed using the Agilent SureSelect XT Human All Exon V6 reagent kit (Agilent Technologies, USA). Bioinformatic analysis was performed as previously described (9). Detected NARS2 variations were further verified using Sanger sequencing (primer sequences available upon request).
In silico analyses
Conservation analysis of NARS2 variants was performed in MegAlign. Potential pathogenicity was predicted using SIFT (http://sift.Jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), MutationTaster (https://www.mutationtaster.org/ChrPos.html), and PROVEAN (http://provean.jcvi.org/index.php). Because NARS2 c.1290G>C changed the first nucleotide of exon 14, the potential effects on mRNA splicing were analyzed using NetGene2 (https://services.healthtech.dtu.dk/service.php?NetGene2-2.42) and Fsplice (http://www.softberry.com/berry.phtml?topic=fsplice&group=programs&subgroup=gfind). Human NARS2 structure was downloaded from SWISS-MODEL (https://swissmodel.expasy.org) as a template (10). Structural modeling of NARS2 variants was performed in RaptorX (http://raptorx.uchicago.edu/StructurePropertyPred/predict/) and SWISS-MODEL (11). All structural figures were managed using the PyMol platform.
mRNA analysis
Peripheral blood mononuclear cells (PBMCs) were isolated from the patient’s father and a healthy control using Ficoll-Paque Plus (GE Healthcare, USA). Total RNA was extracted from PBMCs using the RNeasy Mini Kit (Qiagen, Germany). First-strand cDNA was synthesized using a reverse transcription kit (TaKaRa, China). Synthesized products were amplified with the forward primer, 5'-GAGGCCTCAGAGAAGAACGA-3', located in exon 12 of NARS2 (NM_024678.6), and the reverse primer, 5'-TGGCAAGGGAGAAAAGATGC-3', located in the 3'-untranslated region (3'-UTR) of NARS2. Amplicons were directly sequenced, and those from the father’s cDNA were cloned into the pMD 18-T vector (TaKaRa, China). Eighty-seven clones were selected and sequenced.
Plasmids, cell culture, and transient transfection
Wild-type (WT) NARS2 coding region was synthesized and cloned into pCMV3-FLAG and pCMV3-HA plasmids separately, generating C-terminal FLAG-tagged and C-terminal HA-tagged NARS2. Site-directed mutagenesis was performed using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs, USA). All vectors were confirmed with DNA sequencing. Human embryonic kidney (HEK) 293T cells were obtained from the American Type Culture Collection (ATCC, USA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (v/v) fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37 ℃. Transfection was performed using the jetPRIME reagent (Polyplus Transfection, France), following manufacturer protocol.
Western blot analysis
HEK 293T cells were seeded into 12-well plates and transfected with specific plasmids. Proteins were extracted from whole-cell lysates 24 h post-transfection, separated on 10% SDS-PAGE gels, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with either anti-FLAG (Sigma-Aldrich Cat# F1804, RRID:AB_262044, USA) or anti-GAPDH antibody (Proteintech, Cat# 60004-1-Ig, RRID:AB_2107436, USA). Each analysis was replicated at least three times. Images were analyzed and quantified in ImageJ (12).
Immunofluorescence staining
HEK 293T cells were transfected with WT and mutant FLAG-tagged NARS2 constructs. At 24 h post-transfection, cells were stained with 500 nM MitoTracker Deep Red FM (YEASEN Biotech, China) for 30 min, fixed in 4% paraformaldehyde, permeabilized in 0.25% Triton X-100 in PBS, and blocked in 1% BSA. Immunostaining was performed using mouse anti-FLAG monoclonal antibody (Sigma-Aldrich Cat# F1804, RRID:AB_262044, USA) for 1 h, followed by labelling with Alexa Fluor® 555 conjugated anti-mouse immunoglobulin G (IgG; Cell Signaling Technology Cat# 4409, RRID:AB_1904022, USA) for 1 h. Coverslips were placed on the slides using mounting medium containing DAPI (Thermo Fisher Scientific, USA) and analyzed under a Leica DM6000 fluorescence microscope (Leica Microsystems, Germany). The experiment was replicated three times.
Co-immunoprecipitation
FLAG-tagged NARS2 plasmids, together with HA-tagged NARS2 plasmids, were co-transfected into HEK 293T cells. At 36 h post-transfection, cells were lysed and incubated with anti-HA magnetic beads (Thermo Fisher Scientific Cat# 88836, RRID:AB_2749815, USA) to immunoprecipitate HA-tagged NARS2, following manufacturer protocol. Cell lysates and immune complexes were separated on 10% SDS-PAGE gels, transferred to PVDF membranes, and probed with either anti-FLAG (Sigma-Aldrich Cat# F1804, RRID:AB_262044, USA) or anti-HA antibody (Cell Signaling Technology Cat# 2367, RRID:AB_10691311). This experiment was replicated three times.
Statistical analysis
GraphPad Prism 7.00 version was used for statistical analysis, and the data were expressed as mean ± standard deviation (SD). T-tests were used for comparisons. P<0.05 was considered statistically significant.
Results
Identification of two novel NARS2 variants
Bioinformatic analysis of the WES data showed that the coverage of target region reached 99.8%. The average sequencing depth was 204×, 98.8% of target region reached over 20×. Using WES, we identified two heterozygous variants in NARS2: NARS2 (NM_024678.6) c.1141A>G/p.Asn381Asp and NARS2 (NM_024678.6) c.1290G>C/p.Trp430Cys (Figure 1A).These variants were further confirmed by Sanger sequencing in the pedigree. NARS2 c.1141A>G was maternally inherited, whereas NARS2 c.1290G>C was paternally inherited. After searching through the Human Genome Mutation Database (http://www.hgmd.cf.ac.uk), Genome Aggregation Database (http://gnomad.broadinstitute.org/), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), we concluded that the variants were novel. Our local database (in individuals of the same ethnicity) also did not contain allelic frequencies of the two variants. All the reported pathogenic variants in NARS2 were summarized (Figure 1B).
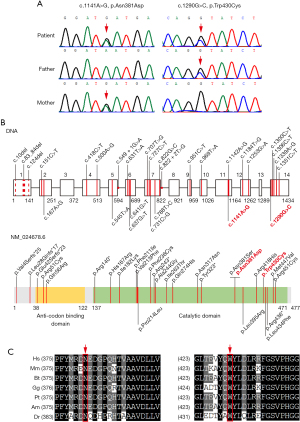
In silico analyses of the NARS2 c.1141A>G and c.1290G>C variants
The NARS2 Asn381 and Trp430 amino acid residues are highly conserved across different species (Figure 1C). Multiple in silico predictive algorithms predicted that NARS2 c.1141A>G/p.Asn381Asp and c.1290G>C/p.Trp430Cys would have deleterious effects, including SIFT (damaging), PolyPhen-2 (probably damaging), MutationTaster (disease-causing), and PROVEAN (damaging). Because NARS2 c.1290G is the first nucleotide of exon 14, we used NetGene2 and Fsplice to assess its effect on NARS2 splicing. NetGene2 showed that the native and variant acceptor sites had a confidence of 0.41 and 0.25, respectively, while Fsplice indicated that their matrix weights were 7.2 and 5.2.
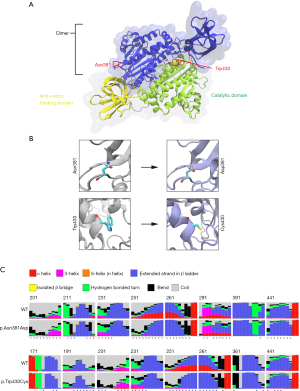
The 3D structure of the NARS2 protein was downloaded from SWISS-MODEL (Figure 2A). NARS2 p.Asn381Asp was located close to the active site, causing the replacement of a polar uncharged hydrophilic amino acid with an acidic amino acid; this change likely influenced protein structure and interactions with nearby areas (Figure 2B). The RaptorX prediction of protein secondary structure revealed significant differences between WT and p.Asn381Asp variants (Figure 2C). NARS2 p.Trp430Cys was also close to the active site, with the variant causing a substitution of the tryptophan indole ring. This change alters a nonpolar amino acid to a polar uncharged amino acid, potentially affecting how the active site binds with key molecules and its catalytic function (Figure 2B). RaptorX again revealed significant changes in the secondary structure (Figure 2C).
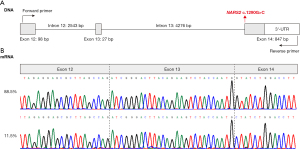
mRNA analysis of NARS2 c.1290G>C
The mRNA analysis detected a band of 638 bp in samples from the patient’s father and the healthy control, but no other bands. Sanger sequencing of the amplicons revealed normal splicing of NARS2 in both the father and the control. After cloning and sequencing of amplicons from the father, we did not detect aberrant transcripts. However, the transcript ratio between NARS2 c.1290G>C and WT was abnormal. Most transcripts were from the NARS2 WT allele, while only 11.5% (10/87) of all sequencing clones were from the NARS2 c.1290G>C allele (Figure 3).
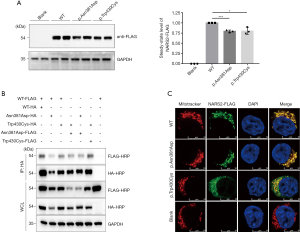
In vitro experiment on NARS2 variants
Western blots indicated that the expression levels of both NARS2 p.Asn381Asp and p.Trp430Cys variants were significantly lower than WT expression (Figure 4A). Also, as we expected, WT NARS2 was transported into the mitochondria. Immunofluorescence staining demonstrated that both p.Asn381Asp and p.Trp430Cys variants also localized to mitochondria (Figure 4B). To analyze whether NARS2 variants affected dimerization, we co-expressed different combinations of FLAG-tagged and HA-tagged NARS2 plasmids in HEK 293T cells. The results of co-immunoprecipitation indicated that both p.Asn381Asp and p.Trp430Cys variants retained their ability to form dimers (Figure 4C).
Discussion
Here, we examined a patient with compound heterozygous variants of NARS2. The patient mainly presented early onset generalized epilepsy, myoclonic seizures, and severe bilateral hearing impairment, which was consistent with the characteristics of COXPD24. He could not lift his head up stably at 3 months old, indicating early developmental delay. Laboratory test suggested affected liver and cardiac function. However, we were not able to further assess his development and heart function, because the patient died of a seizure at 6 months and no cardiological imaging evaluation was performed before death. The patient’s brain MRI was normal at 3 months. No more brain MRI were performed before his death. Possibly more symptoms were masked by his early death.
Nearly 20 patients carrying NARS2 variants have been described since the first variant was reported in 2015 (5-8,13-17). The resultant deleterious phenotypes vary, with well-known examples being Alpers syndrome, Leigh syndrome, mitochondrial myopathy, hemiconvulsion-hemiplegia-epilepsy syndrome, COXPD24, and nonsyndromic deafness. Disease onset in most patients occurs during infancy and involves mitochondrial encephalopathy. However, some reports have also described late-onset patients in their 20s with mild spasticity and weakness or solo non-progressive manifestations, such as hearing loss (8,17). The most common clinical manifestations of NARS2 variants are refractory epilepsy, sensorineural hearing impairment, profound global developmental delay, and hypotonia. Consistent with previously reported cases, our patient exhibited early-onset generalized epilepsy, myoclonic seizures, and severe bilateral hearing impairment. We also noticed persistent elevation of serum hepatic and myocardial enzymes, indicating effects on the liver and heart. Although liver dysfunction has been observed in patients with NARS2 variants, cardiac dysfunction has not (15). Our work here suggests that further investigation is necessary to verify whether NARS2 variants lead to cardiomyopathy.
NARS2 contains an anti-codon binding domain at the N-terminus and a catalytic domain at the C-terminus (18). To date, over 30 NARS2 variants have been identified, including missense, nonsense, splicing variants, and gross deletions (Figure 1B), with missense being the most prevalent. Our study identified two novel NARS2 variants: NARS2 c.1141A>G and NARS2 c.1290G>C. Both were located in the catalytic domain and affected highly conserved amino acid residues. The location is typical of NARS2 variants, although variants can be found throughout the protein. Molecular modeling data suggested that both novel variants significantly changed the secondary structure. Additionally, separate pathogenicity-prediction programs all found them to be pathogenic.
NARS2 c.1141A>G leads to the substitution of asparagine with aspartic acid at position 381 (p.Asn381Asp). In a patient with Leigh syndrome and auditory neuropathy, a different missense change at the same amino acid residue (p.Asn381Ser) was previously in a compound heterozygous state with variant c.969T>A/p.Tyr323* (8). The p.Asn381Ser variant appears to exert a negative effect through disrupting NARS2 dimerization ability, although p.Asn381 is not directly involved in the dimer interphase (8). In addition, p.Asn381Ser decreased NARS2 protein levels in patient fibroblasts (8). We thus performed in vitro experiments to assess the impact of p.Asn381Asp on NARS2 expression and dimerization. Western blot analysis showed that the NARS2 protein with p.Asn381Asp had lower steady-state levels than WT. However, NARS2 dimerization and mitochondrial localization were unaffected.
Single-base substitutions at the first nucleotide position in an exon may affect splicing patterns (19). NARS2 c.1290G is the first nucleotide in the last exon of NARS2. Therefore, we used in silico and mRNA analyses to assess whether NARS2 c.1290G>C causes a splicing defect. Although the c.1290G>C variant acceptor site exhibited a lower splicing score, we did not identify any aberrant splicing in RNA extracted from the patient’s father, who carried the variant. However, we observed reduced transcription of the NARS2 c.1290G>C allele, suggesting that the transcript resulting from this variant was unstable. In vitro expression study also revealed decreased protein levels of NARS2 c.1290G>C/p.Trp430Cys. Similar to NARS2 c.1141A>G, the NARS2 c.1290G>C variant did not affect NARS2 mitochondrial localization and dimerization. The next step is to examine the functional impact of NARS2 variants on the mitochondrial respiratory chain using tissue samples from the patient, given that this link had been proposed in earlier research (6-8,13-15). However, we were unable to do so in this study because no patient tissue was available.
In conclusion, we successfully identified two novel NARS2 variants in a patient with COXPD24, characterized by early-onset generalized epilepsy, myoclonic seizures, and severe bilateral hearing impairment. Both variants impaired NARS2 expression, but did not affect the protein’s mitochondrial localization or dimerization. More experiments are needed to confirm the impact of NARS2 variants on aminoacylation activities and mitochondrial protein synthesis. Nevertheless, our study broadens the clinical and variation spectrum of NARS2-related disorders.
Acknowledgments
Funding: The study was supported by the Clinical Research Plan of SHDC (No. SHDC2020CR3042B to JW), Project of Shanghai Municipal Science and Technology Commission (No. 20MC1920400), and Project of Shanghai Municipal Science and Technology Commission (No. 20dz2260900).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-21-570/rc
Data Sharing Statement: https://tp.amegroups.com/article/view/10.21037/tp-21-570/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-21-570/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-21-570/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of SCMC (No. SCMCIRB-K2020060-1). Informed consent was obtained from the patient’s parents prior to the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bonnefond L, Fender A, Rudinger-Thirion J, et al. Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry 2005;44:4805-16. [Crossref] [PubMed]
- Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet 2009;55:1-18. [Crossref] [PubMed]
- Sissler M, González-Serrano LE, Westhof E. Recent Advances in Mitochondrial Aminoacyl-tRNA Synthetases and Disease. Trends Mol Med 2017;23:693-708. [Crossref] [PubMed]
- Schwenzer H, Zoll J, Florentz C, et al. Pathogenic implications of human mitochondrial aminoacyl-tRNA synthetases. Top Curr Chem 2014;344:247-92. [Crossref] [PubMed]
- Mizuguchi T, Nakashima M, Kato M, et al. PARS2 and NARS2 mutations in infantile-onset neurodegenerative disorder. J Hum Genet 2017;62:525-9. [Crossref] [PubMed]
- Seaver LH, DeRoos S, Andersen NJ, et al. Lethal NARS2-Related Disorder Associated With Rapidly Progressive Intractable Epilepsy and Global Brain Atrophy. Pediatr Neurol 2018;89:26-30. [Crossref] [PubMed]
- Sofou K, Kollberg G, Holmström M, et al. Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Mol Genet Genomic Med 2015;3:59-68. [Crossref] [PubMed]
- Simon M, Richard EM, Wang X, et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet 2015;11:e1005097. [Crossref] [PubMed]
- Yu T, Li J, Li N, et al. Obesity and developmental delay in a patient with uniparental disomy of chromosome 2. Int J Obes (Lond) 2016;40:1935-41. [Crossref] [PubMed]
- Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296-303. [Crossref] [PubMed]
- Källberg M, Wang H, Wang S, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 2012;7:1511-22. [Crossref] [PubMed]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671-5. [Crossref] [PubMed]
- Vanlander AV, Menten B, Smet J, et al. Two siblings with homozygous pathogenic splice-site variant in mitochondrial asparaginyl-tRNA synthetase (NARS2). Hum Mutat 2015;36:222-31. [Crossref] [PubMed]
- Štěrbová K, Vlčková M, Hansíková H, et al. Novel variants in the NARS2 gene as a cause of infantile-onset severe epilepsy leading to fatal refractory status epilepticus: case study and literature review. Neurogenetics 2021;22:359-64. [Crossref] [PubMed]
- Sofou K, Kollberg G, Hedberg-Oldfors C, et al. The phenotypic variability and natural history of NARS2 associated disease. Eur J Paediatr Neurol 2021;31:31-7. [Crossref] [PubMed]
- Vafaee-Shahi M, Farhadi M, Razmara E, et al. Novel phenotype and genotype spectrum of NARS2 and literature review of previous mutations. Ir J Med Sci 2021; Epub ahead of print. [Crossref] [PubMed]
- Souza PVS, Bortholin T, Dias RB, et al. New genetic causes for complex hereditary spastic paraplegia. J Neurol Sci 2017;379:283-92. [Crossref] [PubMed]
- Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med 2013;5:332-43. [Crossref] [PubMed]
- Fu Y, Masuda A, Ito M, et al. AG-dependent 3'-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res 2011;39:4396-404. [Crossref] [PubMed]


