
Whole-exome sequencing establishes a diagnosis of Alstrom syndrome: a case report
Introduction
Alstrom syndrome (ALMS) is a very rare autosomal recessive disease that is characterized by multiple organ involvement and is classified into ciliopathy groups (1). The disease was reported in a large Swedish family in 1959 for the first time (2). Alstrom described a new syndrome with a series of manifestations, including obesity, retinal degeneration, progressive sensorineural deafness, cardiomyopathy and insulin-resistant diabetes. The estimated incidence of ALMS is 1~9 cases per 1 million (3), and nearly 950 cases have been reported to date.
The ALMS1 gene is located on chromosome 2p13 (4), and more than 300 mutations in ALMS1 have been identified. Almost all of the mutations are frameshift or nonsense mutations. The function of the ALMS1 gene is not well understood, but it has been implicated in cell cycle regulation and intracellular transport, in addition to ciliary disease (5). The clinical findings frequently observed in patients with ALMS are sophisticated and vary considerably. The main clinical features include photoreceptor dystrophy, nystagmus, and sensorineural hearing loss (6). Truncal obesity, diabetes mellitus (DM) and dilated cardiomyopathy are also very common manifestations. Other features include acanthosis nigricans, hypertriglyceridemia, and endocrine anomalies.
Here, we present a patient with heterozygous mutations c.8366delT, p.L2789* and c.6829C>T, p.R2277* in ALMS1. c. 8366delT was a novel mutation in ALMS1. The variability among the patient and her affected sisters within the same family is described here. We present the following article in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-21-623/rc).
Case presentation
The patient was an 8-year-and-11-month-old female. She was admitted to the outpatient department because of a one-month-long history of DM with unstable blood glucose levels. Her height was 135 cm, her weight was 46.6 kg, and her BMI was 25.56 kg/m2 (SDS: 2.19). Acanthosis nigricans was noted around the neck and axilla. She had nystagmus in both eyes. She was born after an uneventful pregnancy and delivery (birth parameters: weight, 2,350 g; length, 48 cm). Psychomotor development was normal with attainment of milestones at the appropriate ages. No adverse family history was provided. Ophthalmic tests were not performed due to a lack of cooperation, although photophobia was observed shortly after birth. Her weight gain was significant from 9 months. At 11 months, she underwent a routine physical examination and had fatty liver. Due to her obesity, she had fasting blood glucose and insulin tests, but no abnormalities were found. Her alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels have been mildly elevated since then. Visual disturbance and photophobia developed gradually during the patient’s first year of life. She had one episode of otitis media at 6 years old but had normal hearing. One month before admission, she developed polyuria and weight loss, and she was diagnosed with type 2 DM (T2DM). At a local hospital, twice-daily administration of insulin showed that her blood glucose was unstable. She was then transferred to our hospital.
Laboratory examination revealed ALT was 163.0 (0–40.0) U/L, AST was 118.0 (0–40.0) U/L, creatine was 48.0 (32.3–54.2) µmol/L, blood urea nitrogen was 6.71 (1.8–6.4) mmol/L, triglycerides were 8.34 (0–1.69) mmol/L, low-density lipoproteins were 1.67 (1.25–3.3) mmol/L, and electrolytes were normal. The blood glucose levels fluctuated between 5 and 18 mmol/L. Blood gas analysis showed a pH of 7.36, PCO2 was 29.6 mmHg, HCO3 was 20 mmol/L, and BE was −4 mmol/L. Plasma lactate was 1.8 mmol/L (normal <2); serum ammonia was 52.2 mmol/L (µmol/L) (normal <80). Serum insulin levels were 73.1 (2.6–24.9) U/L (before insulin was administered), HbA1c was 11.4% (4.0–6.0%), and C peptide was 12.1 (0.71–4.22) ng/mL. Glutamate decarboxylase antibody (GAD), zinc finger protein 8 antibody (ZnT8) and human protein tyrosine phosphatase antibody (IA-2A) were negative. Serum-free thyroxine was 12 µg/dL (normal: range, 10.8–20 µg/dL), and insulin growth factor 1 (IGF-1) was 212 (57.0–316.0) ng/mL. Electroencephalogram (EEG) results did not show any abnormalities. Hepatitis A, B, C, D and E antibodies were negative. Epstein-Barr virus DNA was negative. Echocardiography showed no evidence of congenital heart disease or cardiomyopathy. Abdominal ultrasound suggested fatty liver. Ophthalmic investigation revealed hyperopia and astigmatism. Visual acuity was very low (less than 1/10 with the Decimal test). There was a hypopigmented appearance to the retina and severe cone-rod dystrophy.
Given the extremely high levels of insulin, C peptide, obesity and acanthosis nigricans, this patient seemed to fulfill the diagnostic criteria for T2DM. However, in addition to diabetes, she had multiorgan involvement. Thus, syndromic diabetes was highly suspected. Her vision dysfunction could not be explained by diabetic complications. After in-depth communication with the parents, they provided the medical history of her two sisters, who died in infancy. She was the third child of the family and the second child of fraternal twins. Her oldest sister passed away at the age of 4 months from cardiomyopathy and cardiac failure (Figure 1). The elder nonidentical twin sister had photophobia and nystagmus shortly after birth and passed away from cardiac failure after pneumonia at the age of 14 months. The disease progression was similar to that of the first child. Because medical records were discarded, detailed records of the children were no longer available. Based on the above history, we performed a genetic test.
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s mother for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Molecular studies
DNA samples obtained from the family were sequenced to identify the causal gene using whole-exome sequencing (WES). DNA was isolated from peripheral blood using a DNA Isolation Kit (Bioteke Corporation, AU1802, Wuxi, China). Genomic DNA samples (1 µg) were fragmented into 200–300-bp portions using a Covaris Acoustic System (Covaris, Woburn, MA, USA). The DNA fragments were then processed by end repair, A-tailing, adaptor ligation and four-cycle precapture polymerase chain reaction amplification, after which all exons and 50 bp in their adjacent introns were captured by a SeqCap EZ Med Exome Enrichment Kit (Roche, Madison, WI, USA). Postcapture amplification and purification were performed on the DNA library, and sequencing was then performed on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA). The raw data that were produced were then filtered and aligned with the human genome reference (hg19) using the Burrows-Wheeler Aligner (BWA; http://bio-bwa.sourceforge.net/), and variants were identified using NextGene V2.3.4 software (Soft genetics, LLC, State College, PA, USA). The data had a 151.24× mean read depth, and approximately 97.95% of the target bases were covered at a 20× average read depth. The filtered variants were then annotated by using NextGene V2.3.4 and the laboratory’s custom scripts to obtain related information, including conservation of nucleotide bases and amino acids, prediction of the biological functions, frequency in normal populations (compared with 1000 Genomes, ExAC, dbSNP database and local specific databases), and the data from HGMD, ClinVar and OMIM. All variants of pathogenicity were interpreted according to American College of Medical Genetics (ACMG) (7) standards and categorized.
Results
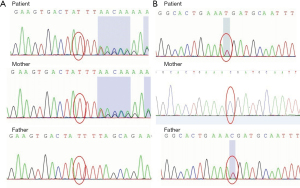
Genetic analysis identified heterozygosity for a frameshift mutation in exon 10 and a nonsense mutation in exon 8 of ALMS1: c.8366delT, p.L2789* and c.6829C>T, p.R2277* (Figure 2). The parents carried each of the variants in a heterozygous state. We have submitted the mutation to the Sequence Read Archive (SRA) database. The reserved accession number is SRR18136620. c.8366delT (PVS1+PM2+PM3) is a novel variant that is predicted to result in the early termination of the protein. c.6829C>T (PVS1+PM2+PM3) has been previously reported in other patients with ALMS.
Discussion
In this study, we report a case of ALMS. The patient has the following characteristics. First, she was found to have nystagmus and photophobia since infancy, and her visual acuity was very low. Retinal dystrophy is a major manifestation that affects 100% of patients with ALMS, and blindness is almost inevitable before the age of 20 (6). Visual symptoms tend to progress slowly but usually occur shortly after birth. The initial assessment should include standard ophthalmic evaluation followed by retinal imaging and functional testing (6). Second, metabolic abnormalities gradually became obvious. Obesity is one of the cardinal features of ALMS (8). ALMS is characterized by subcutaneous fat accumulation rather than in the visceral regions (9). Third, she had hepatic involvement with elevated transaminases from infancy. Given that no virus that commonly causes hepatitis was found, nonalcoholic fatty liver (NAFLD) was probably a reasonable explanation. Gathercole et al. (10) reported an observational cohort study including 30 adult patients with ALMS, and NAFLD and adipose dysfunction were common. Liver biopsies revealed varying degrees of nonalcoholic steatohepatitis (NASH) (inflammation, hepatocyte ballooning, and steatosis).
With increasing age, hypertriglyceridemia, insulin resistance and diabetes had gradually appeared. Because the parents concealed the family history, she was misdiagnosed with T2DM at a local hospital, and all retinopathy was attributed to the complications of T2DM. In fact, when asked carefully, a more detailed medical history was provided. The patient’s original disease manifested first with eye disease and then with diabetes. Based on International Society for Pediatric and Adolescent Diabetes (ISPAD) clinical practice consensus guidelines (11), the diagnosis of T2DM was revised to “genetic syndrome associated with diabetes” after genetic analysis. Unlike insulin resistance caused by simple obesity, ALMS1 mutations affect islet ciliary body function (12).
The onset, progression, and outcome of cardiomyopathy in ALMS are variable. Approximately 70% of patients with ALMS develop congestive heart failure at some point in their lives. Nearly 40% of babies with ALMS develop a transient, severe form of cardiomyopathy by 3 weeks to 4 months of age (8). The proband’s heart was still functioning normally, and her elder sister’s condition seemed to be attributed to infant severe cardiomyopathy. Neither of the proband’s sisters had the opportunity to be genetically screened. However, both elder sisters died of heart failure, and the elder nonidentical twin sister had photophobia and nystagmus shortly after birth. Thus, we deduced that both of them were likely patients as well. Given that the index case did not present any cardiovascular defects but both of her older sisters did, other early-onset conditions, such as dilated cardiomyopathy, infectious myocarditis, muscular dystrophies and in-born errors of metabolism, should also be considered (13). Multiple studies have identified mutations causing cardiomyopathy (14). WES showed no typical mutations in the MYH7 (15), TPM1 (16), TNNT2 (17), MYBPC3 (18), MYL2, MYL3 (19), TNNI3 (20), and ACTC1 (21) genes in this case. Intrafamilial variability in cardiac presentation has been reported previously among four siblings with ALMS (22). The authors reported four siblings who manifested the disease in a variable manner, from severe heart failure to normal systolic function. Intrafamilial variability exists in the onset and degree of severity, ranging from death to spontaneous remission. The manifestation of myocardial involvement was very different among the three children. The mechanism of the variability is unclear. Cardiomyopathy may still appear in adolescence and needs to be followed up.
The differential diagnosis of ALMS includes a series of diseases, such as other monogenetic diabetes, other causes of retinal dystrophies, hereditary cardiomyopathy and Wolfram syndrome. In our patient, Wolfram syndrome in particular needed to be excluded, as it involves abnormalities in the endoplasmic reticulum (ER) and mitochondrial dynamics (23). Wolfram syndrome has similar manifestations to ALMS, such as DM, optic nerve atrophy, sensory deafness and diuresis, and the clinical symptoms range from mild to severe.
We report a novel ALMS1 mutation. c.8366delT is a frameshift mutation that causes the early termination of the protein, p.L2789*. This mutation is located in one of the mutation hotspots of the gene, namely, exon 10. Most pathogenic variants of ALMS1 occur downstream from exon 7, with ‘hotspots’ for disease-causing variants in exons 8, 10, and 16 (24,25). We know that the majority of frameshift variations are predicted to be pathogenic mutations. c.6829C>T was previously reported in Turkish patients with ALMS (26). Our findings extend the genetic spectrum of ALMS1.
No therapy exists to prevent the progressive organ involvement of ALMS. The patient felt well with metformin and insulin, and her blood glucose levels were well under control. Multidisciplinary care is needed.
Conclusions
Identifying the genetic cause of a patient’s diabetes is worthwhile as it directs future monitoring and treatment. It is important to recognize uncommon causes of diabetes, although they are rare. A family history of cardiomyopathy and the manifestation of multisystem involvement in a child may indicate the possibility of ALMS.
Acknowledgments
We thank the family that contributed samples to this study.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-21-623/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-21-623/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s mother for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Álvarez-Satta M, Castro-Sánchez S, Valverde D. Alström syndrome: current perspectives. Appl Clin Genet 2015;8:171-9. [PubMed]
- Alstrom CH, Hallgren B, Nilsson LB, et al. Retinal degeneration combined with obesity, diabetes mellitus and neurogenous deafness: a specific syndrome (not hitherto described) distinct from the Laurence-Moon-Bardet-Biedl syndrome: a clinical, endocrinological and genetic examination based on a large pedigree. Acta Psychiatr Neurol Scand Suppl 1959;129:1-35. [PubMed]
- INSERM. Orphanet: an online rare disease and orphan drug data base. Available online: http://www.orpha.net/
- Collin GB, Marshall JD, Cardon LR, et al. Homozygosity mapping at Alström syndrome to chromosome 2p. Hum Mol Genet 1997;6:213-9. [Crossref] [PubMed]
- Shurygina MF, Parker MA, Schlechter CL, et al. A case report of two siblings with Alstrom syndrome without hearing loss associated with two new ALMS1 variants. BMC Ophthalmol 2019;19:246. [Crossref] [PubMed]
- Tahani N, Maffei P, Dollfus H, et al. Consensus clinical management guidelines for Alström syndrome. Orphanet J Rare Dis 2020;15:253. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Sanyoura M, Woudstra C, Halaby G, et al. A novel ALMS1 splice mutation in a non-obese juvenile-onset insulin-dependent syndromic diabetic patient. Eur J Hum Genet 2014;22:140-3. [Crossref] [PubMed]
- Paisey RB, Hodge D, Williams K. Body fat distribution, serum glucose, lipid and insulin response to meals in Alström syndrome. J Hum Nutr Diet 2008;21:268-74. [Crossref] [PubMed]
- Gathercole LL, Hazlehurst JM, Armstrong MJ, et al. Advanced non-alcoholic fatty liver disease and adipose tissue fibrosis in patients with Alström syndrome. Liver Int 2016;36:1704-12. [Crossref] [PubMed]
- Mayer-Davis EJ, Kahkoska AR, Jefferies C, et al. ISPAD Clinical Practice Consensus Guidelines 2018: Definition, epidemiology, and classification of diabetes in children and adolescents. Pediatr Diabetes 2018;19:7-19. [Crossref] [PubMed]
- Bettini V, Maffei P, Pagano C, et al. The progression from obesity to type 2 diabetes in Alström syndrome. Pediatr Diabetes 2012;13:59-67. [Crossref] [PubMed]
- Lahm H, Jia M, Dreßen M, et al. Congenital heart disease risk loci identified by genome-wide association study in European patients. J Clin Invest 2021;131:141837. [Crossref] [PubMed]
- Burke MA, Cook SA, Seidman JG, et al. Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. J Am Coll Cardiol 2016;68:2871-86. [Crossref] [PubMed]
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell 1990;62:999-1006. [Crossref] [PubMed]
- Thierfelder L, Watkins H, MacRae C, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell 1994;77:701-12. [Crossref] [PubMed]
- Watkins H, McKenna WJ, Thierfelder L, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med 1995;332:1058-64. [Crossref] [PubMed]
- Bonne G, Carrier L, Bercovici J, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet 1995;11:438-40. [Crossref] [PubMed]
- Poetter K, Jiang H, Hassanzadeh S, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet 1996;13:63-9. [Crossref] [PubMed]
- Kimura A, Harada H, Park JE, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet 1997;16:379-82. [Crossref] [PubMed]
- Mogensen J, Klausen IC, Pedersen AK, et al. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest 1999;103:R39-43. [Crossref] [PubMed]
- Hoffman JD, Jacobson Z, Young TL, et al. Familial variable expression of dilated cardiomyopathy in Alström syndrome: a report of four sibs. Am J Med Genet A 2005;135:96-8. [Crossref] [PubMed]
- Samara A, Rahn R, Neyman O, et al. Developmental hypomyelination in Wolfram syndrome: new insights from neuroimaging and gene expression analyses. Orphanet J Rare Dis 2019;14:279. [Crossref] [PubMed]
- Marshall JD, Hinman EG, Collin GB, et al. Spectrum of ALMS1 variants and evaluation of genotype-phenotype correlations in Alström syndrome. Hum Mutat 2007;28:1114-23. [Crossref] [PubMed]
- Marshall JD, Maffei P, Collin GB, et al. Alström syndrome: genetics and clinical overview. Curr Genomics 2011;12:225-35. [Crossref] [PubMed]
- Ozantürk A, Marshall JD, Collin GB, et al. The phenotypic and molecular genetic spectrum of Alström syndrome in 44 Turkish kindreds and a literature review of Alström syndrome in Turkey. J Hum Genet 2015;60:1-9. [Crossref] [PubMed]



