
Bioinformatic analysis of dysregulated circular RNAs in pediatric pulmonary hypertension linked congenital heart disease
Introduction
Pulmonary artery hypertension (PAH) is a term encompassing a cluster of diseases characterized by increased pulmonary vascular resistance. It is a particularly prevalent and critical complication of left-to-right shunt congenital heart disease (CHD), a condition which can lead to right ventricular failure and even sudden death. CHD is the most important cardiovascular disease and the most common birth defect in children. According to surveys conducted in many regions of China over the past decade, the incidence of CHD is approximately 4–22%, indicating it places a serious burden on families and society (1,2). Compared to adults, a low incidence of idiopathic pulmonary arterial hypertension (IPAH) is found in children. Further, approximately 24–52% of pediatric PAH cases are associated with CHD (CHD-PAH), reflecting a higher incidence amongst CHD groups (3,4). Even after complete surgical CHD repair, PAH persistent or recurrent children experience significant pulmonary vascular changes like those with IPAH (5). While its pathogenesis has not yet been fully elucidated, data have shown key genetic risk factors associated with CHD-PAH can lead to a lack of PAH treatment response (6). This leads to rapid disease progression, even when the hemodynamic manifestations are mild or similar. However, the underlying etiology of this phenomenon remains unclear (7,8).
Therefore, in addition to hemodynamic manifestations, it is crucial to study PAH etiology and pathogenesis, as this would aid to reverse pulmonary vascular disease and explore new targets for preventing and treating the condition. While most mammalian genomes are transcribed into RNA, only a small number of said transcripts are translated into proteins. An increasing number of noncoding RNAs have been found that do not encode homologous peptides, yet they still play a major role in the regulation of many basic cellular processes and tissue-specific physiological functions (9). Circular RNAs (circRNAs) comprise a sub-category of these functionally vital, yet noncoding RNAs and are characterized by a covalently closed loop structure, and very stable and not easily degraded expression. CircRNAs share distinct advantages in the development and application of effective treatments for cardiovascular diseases, especially in fields centered on developing novel clinical diagnostic markers and molecular targeted therapies (10,11). In humans, mice, and nematodes, previous reports have shown circRNAs act as microRNA (miRNA) sponges (12), and also participate in the transcriptional regulation of cancer, neurological diseases, and other pathologies (13). PAH and cancer cells show similar high proliferation and anti-apoptotic phenotypes, and studies have confirmed PAH links to differentially expressed circRNAs (14,15). However, the biological functions and regulatory mechanisms of PAH circRNAs remain elusive, and questions surrounding their role in the condition present imperative scientific problems worthy of in-depth research. We applied microarray analysis to identify circRNAs differentially expressed between PAH patients and CHD control subjects, then verified their expression levels in a larger sample. Prediction of miRNAs regulated by circRNAs and their downstream target genes were then developed. Using bioinformatic methods, we can now construct circRNA-related targeted regulatory pathways to attain further insight. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-117/rc).
Methods
Patients and samples
The key aspects of study design (including the research question, primary outcome to be measured and the statistical analysis plan) were prepared and registered before data collection began. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University [No. 2021(KY-E-050)], and parental consent was sought and obtained prior to all research. Eight patients with CHD-PAH (PAH group) and five with PAH-free CHD (the control group) were enrolled in the microarray study, while 16 patients from each group (PAH and control) were enrolled in our validation study. All patients were hospitalized at the Department of Pediatrics from May 2020 to December 2020. PAH was diagnosed by right-heart catheterization based on criteria set as a pre-intervention, with a mean resting pulmonary arterial pressure of ≥25 mmHg. Patients who were diagnosed with other systemic diseases, such as pulmonary dysplasia, arrhythmia, or associated disease, were excluded, as were those who received targeted therapy for PAH. The characteristics of the 13 patients who provided samples for the microarray study are summarized in Table 1. In our validation study, 7 boys and 9 girls were included in the CHD group, with a median age of 4 (3.48–5.38) years, while 9 boys and 7 girls were included in the PAH group, with a median age of 2.75 (0.8–5.48) years.
Table 1
| Patient ID | Group | Gender | Weight (kg) | Age (y) | Diagnosis | CHD diameter (mm) | PAP [mmHg] | |
|---|---|---|---|---|---|---|---|---|
| HYK | PAH | M | 9.2 | 1.3 | VSD | 8.8 | 66/34 [40] | |
| HY | PAH | M | 16.8 | 9.2 | ASD | 39 | 86/53 [64] | |
| LHJ | PAH | F | 11.0 | 4.2 | VSD | 18 | 85/44 [58] | |
| MJQ | PAH | F | 16.4 | 5.8 | VSD | 18 | 73/44 [54] | |
| TWM | PAH | M | 5.6 | 0.4 | VSD | 14 | 66/30 [41] | |
| PBC | PAH | M | 6.3 | 0.58 | VSD | 9 | 68/25 [42] | |
| WLY | PAH | F | 5.2 | 0.8 | VSD | 10 | 90/48 [63] | |
| HXH | PAH | M | 8.3 | 1.8 | * | NA | 85/42 [56] | |
| TBQ | Control | M | 32.4 | 12.8 | VSD | 12 | 27/14 [18] | |
| WYY | Control | M | 12.4 | 3.9 | VSD | 9 | 22/10 [14] | |
| TBY | Control | F | 13.8 | 3.8 | VSD | 5 | 25/10 [15] | |
| ZQY | Control | F | 13.2 | 3.5 | VSD | 5.1 | 21/10 [14] | |
| TYX | Control | M | 14.5 | 3.0 | VSD | 5 | 17/11 [14] | |
| P value | 1.0 | 0.06 | 0.26 | 1.0 | NA | <0.01 | ||
*, ectopic origin of right pulmonary artery from the ascending aorta and PDA. CHD, congenital heart disease; PAP, pulmonary artery pressure; PAH, pulmonary artery hypertension; VSD, ventricular septal defect; ASD, atrial septal defect; PDA, patent ductus arteriosus.
Blood was collected from the femoral vein before cardiac catheterization, and the plasma was separated by centrifugation at 3,000 rpm for 15 min then held at −80 ℃ until total RNA was isolated.
circRNA expression-profile data
The purity and concentration of total RNA was determined using a NanoDrop ND-1000 (Thermo Fisher Scientific, USA). After digestion with RNase R (Epicentre, Inc., USA), the enriched circRNAs were then amplified and transcribed into fluorescent complementary RNA (cRNA) utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar, USA). The labeled cRNAs were hybridized to the Human circRNA Array (8×15 K, Arraystar) and array pictures were then scanned and analyzed using the Agilent software package. R software was used for data analysis and subsequent processing. Differential circRNA expression between the two samples was identified by filtering and supported fold-changes, and distinguishable circRNA expression patterns within the samples were identified by hierarchical clustering. The volcano plot shows known differentially expressed circRNAs. Student’s t-tests were used to calculate statistical significance, while a fold-change of >1.5 with a P value of <0.05 served as our cutoff criterion for significant differential expression.
Reverse transcription and quantitative polymerase chain reaction ((RT-qPCR) validation
Total RNA, including circRNAs, was extracted from the plasma using a BIOG cfRNA Easy Kit (Changzhou Baidai Biotechnology Co., Ltd., China) according to the manufacturer’s instructions. Plasma total RNA was reverse transcribed into complementary DNA (cDNA) using HiScript III RT SuperMix for qPCR (+gDNA wiper) (Vazyme Biotech Co., Ltd., China), and subsequent real-time PCR assays were then performed with plasma samples from PAH patients and CHD controls. All primers were designed and synthesized by Guangzhou Geneseed Biotech (Table 2). The qPCR results were normalized to the reference gene GAPDH. circRNA relative expression levels were then calculated using the 2−∆∆Ct method (16).
Table 2
| Name | Reverse (5′–3′) | Forward (5′–3′) |
|---|---|---|
| hsa_circ_005372 | TTAAACTGCGGATTGCTTGT | CTCTCATCAAGCCTGCATCA |
| hsa_circ_101465 | TCAGGTTCAGGAGCACGGGAAT | TCATGTATGCCCTGGCCTTCGGT |
| hsa_circ_010921 | GCCTGCCAACCTCACCATTC | GCAGATGGGAAAGCGATGGC |
| hsa_circ_003416 | ATTTAAACTTGATCCAACATGC | CCCCTTTCACATCAAAGAAC |
| hsa_circ_008882 | TAATGCTAGGCTGCCAATGGT | GCAGGAATACCTTTCCTCACAG |
circRNA, circular RNA.
Functional analysis and regulative network
circRNA–miRNA and miRNA–messenger RNA (mRNA) interactions were predicted using Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda (17,18). We then studied the miRNA-response elements (MREs) related to qRT-PCR differentially expressed circRNAs to explore the circRNA–miRNA interactions in detail. Target miRNA genes were predicted using Arraystar’s software, then Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to predict the functions of hsa_circ_005372 and hsa_circ_003416. A circRNA–miRNA–mRNA regulatory network was then established using Cytoscape (http://www.cytoscape.org/).
Statistical analysis
All statistical analyses were performed using SPSS version 25.0 (SPSS, USA). Intergroup clinical or demographic variation was determined by either Student’s t-test or Fisher’s exact test, and Mann-Whitney U tests were performed to determine the relative circRNA expression levels. Statistical significance was set at P<0.05, and every applied mathematics test was bilateral.
Results
circRNA profiles in PAH and control samples
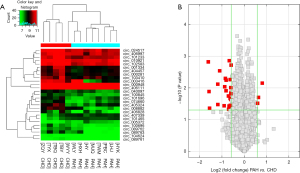
In our systematic study of PAH circRNA expression levels, we performed circRNA microarray analysis of both PAH and control groups. Hierarchical cluster analysis showed the circRNA expression patterns were very different between these groups, as reflected in red and green shading points for high and low respective circRNA expression levels seen in Figure 1A (fold change >1.5, P<0.05). Red dots in the volcano map represent circRNAs that behaved markedly differently between the two samples, with a fold change of >1.5 (Figure 1B). The overall results suggest that 11,611 circRNAs were differentially expressed in PAH samples (fold change ≥1), with three circRNAs significantly upregulated and 24 circRNAs significantly downregulated (fold change ≥1.5, P<0.05).
Validation of dysregulated circRNAs
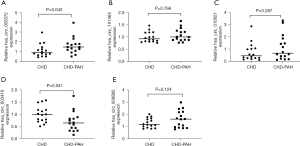
We selected five dysregulated circRNAs to further validate our microarray expression results, including two upregulated circRNAs (005372, 101465) and three downregulated circRNAs (010921, 003416, 008882). To achieve this, we selected based on the following criteria: raw intensity >100, fold change ≥1.5, P<0.05. Samples from 32 patients with CHD, including 16 with CHD-PAH, were included in the validation group, and qRT-PCR was performed to detect the differential expression levels in the patient’s plasma samples. In agreement with the microarray results, hsa_circ_005372 was significantly dysregulated between the case and control groups (Figure 2A). By contrast, hsa_circ_101465, hsa_circ_010921, and did not differ significantly between the two groups (Figure 2B,2C). Hsa_circ_003416 was significantly dysregulated (Figure 2D), while hsa_circ_008882 did not differ significantly between the two groups (Figure 2E).
Prediction of miRNA binding to differentially expressed circRNAs
The miRNAs predicted to be associated with twenty-seven differentially expressed circRNAs are summarized in Table 3. Among them, hsa_circ_407339 and hsa_circ_405324 were associated with the highest number of miRNAs (1,863 and 939, respectively).
Table 3
| Name | Regulation | Number of miRNA targets |
|---|---|---|
| hsa_circ_407339 | Up | 1863 |
| hsa_circ_405324 | Down | 939 |
| hsa_circ_001334 | Down | 903 |
| hsa_circ_000948 | Down | 650 |
| hsa_circ_406111 | Down | 386 |
| hsa_circ_406997 | Down | 346 |
| hsa_circ_089763 | Down | 222 |
| hsa_circ_074660 | Down | 220 |
| hsa_circ_089761 | Down | 192 |
| hsa_circ_404457 | Down | 146 |
| hsa_circ_089762 | Down | 108 |
| hsa_circ_406828 | Down | 97 |
| hsa_circ_040097 | Down | 79 |
| hsa_circ_005372 | Up | 78 |
| hsa_circ_000281 | Down | 78 |
| hsa_circ_010921 | Down | 53 |
| hsa_circ_003416 | Down | 45 |
| hsa_circ_101233 | Down | 43 |
| hsa_circ_104624 | Down | 42 |
| hsa_circ_102583 | Down | 41 |
| hsa_circ_101696 | Down | 33 |
| hsa_circ_008882 | Down | 31 |
| hsa_circ_100696 | Down | 31 |
| hsa_circ_024517 | Down | 30 |
| hsa_circ_100845 | Down | 23 |
| hsa_circ_101465 | Up | 10 |
| hsa_circ_103410 | Down | 8 |
miRNA, microRNA.
MREs that could describe miRNA binding to hsa_circ_005372 and hsa_circ_003416 were also predicted, and the top five MREs predicted for hsa_circ_005372 were miR-548g-3p, miR-365b-5p, miR-365a-5p, miR-216a-5p, and miR-873-5p (Figure 3A). The top five MREs predicted for hsa_circ_003416 were miR-1244, miR-4747-5p, miR-6755-3p, miR-548an, and miR-3194-5p (Figure 3B).

Functional analysis of qRT-PCR-verified deregulated circRNAs
GO function and KEGG enrichment analyses were performed for the two circRNAs showing significant differential expression by qRT-PCR, and we identified 51 GO biological processes (BPs), 10 GO cellular components (CCs), 10 GO molecular functions (MFs), 71 GO functions, and seven KEGG pathways. The differentially expressed circRNAs were related to terms such as cellular response to retinoic acid, cellular protein complex disassembly, platelet alpha granule, cyclin-dependent protein kinase holoenzyme complex, endopeptidase inhibitor activity, and enzyme inhibitor activity (Table 4). Oxidative phosphorylation and tight junction signaling pathways proved the most enriched pathways associated with PAH (Table 5).
Table 4
| GO.ID | Term | P value | Genes |
|---|---|---|---|
| Biological process | |||
| GO: 0071300 | Cellular response to retinoic acid | 0.005 | BRINP3/NDUFA13/SERPINF1 |
| GO: 0043624 | Cellular protein complex disassembly | 0.006 | KIF2A/SYNJ1/VILL/MRPL16/MRPL40 |
| GO: 0010447 | Response to acidic ph | 0.007 | GIP/SERPINF1 |
| GO: 0070830 | Bicellular tight junction assembly | 0.007 | CLDN20/PARD6A/CRB3 |
| GO: 0120192 | Tight junction assembly | 0.008 | CLDN20/PARD6A/CRB3 |
| Cellular component | |||
| GO: 0031091 | Platelet alpha granule | 0.012 | APLP2/TMSB4X/STXBP3 |
| GO: 0000307 | Cyclin-dependent protein kinase holoenzyme complex | 0.022 | CCNH/CCNB3 |
| GO: 0005923 | Bicellular tight junction | 0.028 | CLDN20/PARD6A/CRB3 |
| GO: 0005856 | Cytoskeleton | 0.032 | KIF2A/EIF6/CETN3/SPATC1/PARD6A/CCDC28B/CCDC15/CCNB3/ACTA2/TBCB/SYNJ1/KRTAP131/CCDC39/CTNNA2/VILL/ZBTB49/ODF3L1/RAI14/TMSB4X |
| GO: 0070160 | Tight junction | 0.032 | CLDN20/PARD6A/CRB3 |
| Molecular function | |||
| GO: 0004866 | Endopeptidase inhibitor activity | 0.017 | APLP2/SERPINF1/CST8/SMR3A |
| GO: 0004857 | Enzyme inhibitor activity | 0.017 | PDC/ENSA/SMR3A/APLP2/SERPINF1/CST8 |
| GO: 0030414 | Peptidase inhibitor activity | 0.019 | SMR3A/APLP2/SERPINF1/CST8 |
| GO: 0061135 | Endopeptidase regulator activity | 0.020 | SMR3A/APLP2/SERPINF1/CST8 |
| GO: 0015026 | Coreceptor activity | 0.027 | LRP5/LILRA4 |
GO, Gene Ontology, circRNA, circular RNA.
Table 5
| Pathway ID | Term | P value | Genes |
|---|---|---|---|
| hsa04662 | B cell receptor signaling pathway | 0.006 | IFITM1/LILRA1/LILRA4 |
| hsa03420 | Nucleotide excision repair | 0.020 | CCNH/LIG1 |
| hsa00190 | Oxidative phosphorylation | 0.023 | ATP6V1H/COX6A1/NDUFA13 |
| hsa05226 | Gastric cancer | 0.031 | CTNNA2/FGF3/LRP5 |
| hsa05131 | Shigellosis | 0.038 | DOCK1/U2AF1L4 |
| hsa04530 | Tight junction | 0.043 | CLDN20/CRB3/PARD6A |
| hsa05100 | Bacterial invasion of epithelial cells | 0.045 | CTNNA2/DOCK1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; circRNA, circular RNA.
Construction of a circRNA–miRNA–mRNA regulative network
Having predicted MREs and targeted genes, and as informed by qRT-PCR results confirming very different circRNA expression levels, we built a regulative interaction map of PAH-related circRNA–miRNA–mRNA. This schematic included 93 mRNAs and 29 miRNAs for hsa_circ_005372, and 26 mRNAs and 19 miRNAs for hsa_circ_003416 (Figure 4).

Discussion
Vascular endothelial functionality disorders link to a chronic state of high pulmonary blood flow. In turn, this leads to imbalances in various vascular contraction and diastolic factors between collagen generation and degradation, causing pulmonary vascular remodeling which eventually leads to PAH in left-to-right shunt CHD patients (19). Nonetheless, a PAH pathogenic mechanism has yet to be completely elucidated. Other studies have shown that the occurrence of CHD in PAH patients cannot be completely explained by hemodynamics, genetic mutations, or clinical phenotype related epigenetic modification (20-22), and mounting evidence has indicated that wide ranges of circRNAs are abnormally expressed in PAH (23-25). However, the specific involvement of circRNAs in CHD-PAH pathogenesis merits further study. For the first time, this study analyzed plasma circRNA expression levels in children with CHD-PAH, and identified numerous circRNAs that were aberrantly expressed compared with children with PAH-free CHD.
We identified and analyzed circRNA plasma levels in PAH patients with CHD to further our efforts to explore their potential pathogenic roles. As a result, we found certain circRNAs were differentially expressed in PAH, and two were the most significantly dysregulated in children with the condition. Application of GO and KEGG analyses then revealed BPs and signaling pathways linked to both of these differentially expressed circRNAs. Bioinformatic analysis indicated that both might be related to GO phenomena, including endopeptidase inhibitor activity, enzyme inhibitor activity, platelet alpha granule, cyclin-dependent protein kinase holoenzyme complex, cellular response to retinoic acid, and cellular protein complex disassembly, while KEGG analysis suggested the oxidative phosphorylation and tight junction signaling pathways were significantly enriched. This is likely related to PAH occurrence and regulation processes as mentioned in previous studies (26,27). Abnormal regulation of oxidative signaling is closely related to cell apoptosis, and antioxidant therapy has been shown to have a certain effect on PAH (28). Changes in tight junction signaling may affect cell proliferation and migration (29), while the proliferation, migration, and apoptosis of pulmonary smooth muscle cells (PASMCs) are the main pulmonary vascular remodeling changes found in PAH (30). Therefore, these two abnormally regulated circRNAs and their related signaling pathways are expected to become new therapeutic targets for PAH, although the mechanics informing their regulation of PASMC proliferation, migration, and apoptosis merits further study.
In recent years, the mechanisms of different PAH circRNAs have been reported. In a previous study, serum hsa_circ_0029642 expression was significantly lower in patients with CHD-PAH compared to patients with normal pulmonary artery pressure, suggesting it could serve as a new serum marker for PAH (31), and similar results have been observed in other types of PAH. Wang et al. (24) performed circRNA microarray and bioinformatic analyses on lung tissues from hypoxic pulmonary hypertensive mice and found several potentially diagnostic and therapeutic circRNA targets for the condition. Further experimentation revealed circRNAs are related to vascular remodeling and PAH pathogenesis by affecting the proliferation of miRNA promoted PASMCs (32,33). In another study, Miao et al. (14) analyzed the circRNA profile and identified hsa_circ_0002062 and hsa_circ_0022342 as potential therapeutic targets in the peripheral blood of chronic thromboembolic pulmonary hypertensive patients. Similarly, Zhang (34) found serum circ_0068481 levels were elevated in patients with IPAH, suggesting it could be used to diagnose the condition, predict adverse clinical outcomes, and serve as a novel and non-invasive biomarker. Although the etiologies are different, with manifestations such as hypoxia and thromboembolism, circRNAs play important roles in the various types of PAH. The common molecular mechanisms that are evident in PAH are also reflected in its genetic etiology. In turn, identically variant gene expression was found across patients with CHD-PAH, connective tissue disease PAH, and IPAH (35). Further, in our present study, hsa_circ_005372 and hsa_circ_003416 were significantly dysregulated and may play a role in regulating the expression of both host genes and downstream target genes. However, questions as to whether these factors can be used as potential diagnostic biomarkers or therapeutic targets still merit further testing on larger samples.
The circRNAs, similar to long non-coding RNAs and mRNAs, contain many miRNA-binding sites and function through a variety of mechanisms. These mechanisms include acting as competitive endogenous RNAs (ceRNAs), regulating gene transcription, directly interacting with proteins, and influencing protein translation (36). The miRNA sponge-like functions of circRNAs, as well as their ability to regulate downstream miRNA-target gene expression as ceRNAs, comprise the most widely studied of these mechanisms (37). In one study, hsa_circ_0016070 was shown to be related to vascular remodeling and to participate in PAH pathogenesis by influencing miR-942/CCND1 to promote pulmonary artery smooth muscle cell proliferation (38). In 2020, Miao et al. (39) predicted that, based on an analysis of peripheral blood samples, the hsa_circ_0046159–miR-1226-3p–ATP2A2 pathway was linked to CTEPH progression.
To discern the specific biological consequences of the two abnormally expressed circRNAs, a network including circRNA–miRNA–mRNA interaction was constructed and revealed possible connections between circRNAs and their target genes. In addition, the network served as an important study reference on the regulatory mechanisms between circRNAs and their potential downstream targets. We observed that hsa_circ_005372 and hsa_circ_003416 could bind to many miRNAs, some of which have been associated with cell proliferation, migration, and differentiation. For example, miR-3125 (40), miR-216a-5p (41), and miR-365a-5p (42) were identified as a novel upregulated circRNA’s targets (hsa_circ_005372) while, by contrast, miR-199a/b-3p, miR-3194-5p (43), and miR-486-3p (44) were identified as targets of a novel downregulated form of circRNA (hsa_ circ_003416). Among these, miR-199a-3p was previously linked to PAH pathogenesis, which was indicated by the prediction of potential hub gene targeting drugs (45,46), lending new insights into future therapies. Potential mRNA targets for miR-199a-3p include TMSB4X (47) and ACTA2 (48), which are related to vasoconstriction and cell apoptosis and play key roles in PAH. While circRNAs and miRNAs may target many genes, resulting in changes in many downstream targets, a comprehensive understanding of their exact biogenesis and regulatory circuits is lacking at present (49). Research on the roles of circRNAs in the occurrence and regulation of PAHs is in its infancy, and further exploration is needed and anticipated.
The detection of circRNAs related markers is expected to provide a noninvasive detection method for the diagnosis, prognosis and treatment of PAH. However, the research of circRNAs in PAH is still in its infancy. There are still many challenges: first, the expression level of most circRNAs is low, and more advanced and sensitive technologies and tools need to be developed to detect the quantification and verification of circRNAs; Second, the sequence of most circRNA is synchronized with the mRNA produced by the host gene, so it also faces technical challenges, such as overexpression and silencing strategies (12). At the same time, the mechanisms of circRNA biogenesis are still fairly elusive. Current studies suggest that the biogenesis of circRNA is regulated by specific cis-acting elements and trans-acting factors (50,51). And a number of RNA-binding proteins regulate and control circRNA biogenesis. Gene body methylation also plays a regulatory role in circRNA biogenesis. In addition, epigenetic changes within histones and gene bodies affect alternative splicing and may also have a direct impact on circRNAs biogenesis. Exploring the factors that affect biogenesis will help us understand their function better. The biogenesis and exact function of circRNA and its function and mechanism in different human diseases are still unclear. Therefore, further research is needed in the future to make circRNA play a greater role in the prevention, diagnosis and treatment of PAH and realize clinical transformation faster.
One limitation of this study is that we only verified the differential expression of circRNAs in 16 pairs of patients, and a larger sample size is needed to confirm these results and long-term follow-up and further evaluation of circRNAs expression changes after treatment are required to develop biomarker circRNAs. Moreover, the specific function of these abnormally expressed PAH circRNAs and the degree to which they are involved in PAH pathogenesis still awaits cellular and animal model confirmation. To date, limited research has been conducted on the interactions between PAH circRNAs and miRNAs, and the results of our study have established a circRNA–miRNA–gene PAH network which will inform further explorations of the roles circRNAs play in PAH pathogenesis.
In conclusion, we identified differentially expressed PAH circRNAs and showed their dysregulation may link to PAH pathogenesis. The results of this study can broaden perspectives on PAH epigenetic research and lay a preliminary foundation for future studies on the roles of PAH circRNAs.
Acknowledgments
The authors thank all participating families and patients for their contributions. The authors would also like to thank all physicians at The First Affiliated Hospital of Guangxi Medical University, Department of Pediatrics-Division I, for their help and support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-117/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-117/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-117/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to its accuracy and integrity are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University [No. 2021(KY-E-050)]. Written informed consent was obtained from the parents of all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li SL, Gu RY, Huang GY. Epidemiological features of congenital heart disease in Chinese children. Chinese Journal of Practical Pediatrics 2017;32:871-5.
- Hu SS. Commentary on series of Chinese expert consensus on surgical treatment of congenital heart diseases. Chinese Journal of Clinical Thoracic and Cardiovascular Surgery 2021;28:1-3.
- Wu DC, Zhang HD, Jing ZC. Pediatric pulmonary arterial hypertension. Curr Hypertens Rep 2013;15:606-13. [Crossref] [PubMed]
- Kwiatkowska J, Zuk M, Migdal A, et al. Children and Adolescents with Pulmonary Arterial Hypertension: Baseline and Follow-Up Data from the Polish Registry of Pulmonary Hypertension (BNP-PL). J Clin Med 2020;9:1717. [Crossref] [PubMed]
- Latus H, Wagner I, Ostermayer S, et al. Hemodynamic Evaluation of Children with Persistent or Recurrent Pulmonary Arterial Hypertension Following Complete Repair of Congenital Heart Disease. Pediatr Cardiol 2017;38:1342-9. [Crossref] [PubMed]
- Girerd B, Lau E, Montani D, et al. Genetics of pulmonary hypertension in the clinic. Curr Opin Pulm Med 2017;23:386-91. [Crossref] [PubMed]
- Pattathu J, Gorenflo M, Hilgendorff A, et al. Genetic testing and blood biomarkers in paediatric pulmonary hypertension. Expert consensus statement on the diagnosis and treatment of paediatric pulmonary hypertension. The European Paediatric Pulmonary Vascular Disease Network, endorsed by ISHLT and DGPK. Heart 2016;102:ii36-41. [Crossref] [PubMed]
- Yuan SM. Pulmonary artery hypertension in childhood: The transforming growth factor-β superfamily-related genes. Pediatr Neonatol 2018;59:112-9. [Crossref] [PubMed]
- Bonnet S, Boucherat O, Paulin R, et al. Clinical value of non-coding RNAs in cardiovascular, pulmonary, and muscle diseases. Am J Physiol Cell Physiol 2020;318:C1-C28. [Crossref] [PubMed]
- Tang Y, Bao J, Hu J, et al. Circular RNA in cardiovascular disease: Expression, mechanisms and clinical prospects. J Cell Mol Med 2021;25:1817-24. [Crossref] [PubMed]
- Zhu L, Li N, Sun L, et al. Non-coding RNAs: The key detectors and regulators in cardiovascular disease. Genomics 2021;113:1233-46. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Mao L, Guo J, Hu L, et al. Circular RNAs in childhood-related diseases and cancers: A review. Cell Biochem Funct 2021;39:458-67. [Crossref] [PubMed]
- Miao R, Wang Y, Wan J, et al. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine (Baltimore) 2017;96:e7354. [Crossref] [PubMed]
- Xu SL, Deng YS, Liu J, et al. Regulation of circular RNAs act as ceRNA in a hypoxic pulmonary hypertension rat model. Genomics 2021;113:11-9. [Crossref] [PubMed]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101-8. [Crossref] [PubMed]
- Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol 2003;5:R1. [Crossref] [PubMed]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012;13:271-82. [Crossref] [PubMed]
- Oishi P, Fineman JR. Pulmonary Hypertension. Pediatr Crit Care Med 2016;17:S140-5. [Crossref] [PubMed]
- Chen W, Li S. Circulating microRNA as a Novel Biomarker for Pulmonary Arterial Hypertension Due to Congenital Heart Disease. Pediatr Cardiol 2017;38:86-94. [Crossref] [PubMed]
- Ma K, Zhao Q, Chen W, et al. Human lung microRNA profiling in pulmonary arterial hypertension secondary to congenital heart defect. Pediatr Pulmonol 2015;50:1214-23. [Crossref] [PubMed]
- Cheng X, Wang Y, Du L. Epigenetic Modulation in the Initiation and Progression of Pulmonary Hypertension. Hypertension 2019;74:733-9. [Crossref] [PubMed]
- Wang J, Zhu M, Pan J, et al. Circular RNAs: a rising star in respiratory diseases. Respir Res 2019;20:3. [Crossref] [PubMed]
- Wang J, Zhu MC, Kalionis B, et al. Characteristics of circular RNA expression in lung tissues from mice with hypoxia-induced pulmonary hypertension. Int J Mol Med 2018;42:1353-66. [Crossref] [PubMed]
- Jin X, Xu Y, Guo M, et al. hsa_circNFXL1_009 modulates apoptosis, proliferation, migration, and potassium channel activation in pulmonary hypertension. Mol Ther Nucleic Acids 2021;23:1007-19. [Crossref] [PubMed]
- Sithamparanathan S, Rocha MC, Parikh JD, et al. Skeletal muscle mitochondrial oxidative phosphorylation function in idiopathic pulmonary arterial hypertension: in vivo and in vitro study. Pulm Circ 2018;8:2045894018768290. [Crossref] [PubMed]
- Jandl K, Marsh LM, Hoffmann J, et al. Basement Membrane Remodeling Controls Endothelial Function in Idiopathic Pulmonary Arterial Hypertension. Am J Respir Cell Mol Biol 2020;63:104-17. [Crossref] [PubMed]
- Hansen T, Galougahi KK, Celermajer D, et al. Oxidative and nitrosative signalling in pulmonary arterial hypertension - Implications for development of novel therapies. Pharmacol Ther 2016;165:50-62. [Crossref] [PubMed]
- Díaz-Coránguez M, Liu X, Antonetti DA. Tight Junctions in Cell Proliferation. Int J Mol Sci 2019;20:5972. [Crossref] [PubMed]
- He S, Zhu T, Fang Z. The Role and Regulation of Pulmonary Artery Smooth Muscle Cells in Pulmonary Hypertension. Int J Hypertens 2020;2020:1478291. [Crossref] [PubMed]
- Peng WL, Zhao J, Chao L, et al. Correlation of circular RNA hsa_circ_0029642 with pulmonary hypertension in adult patients with congenital heart disease. Journal of Army Medical University 2017;39:1005-8.
- Ma C, Gu R, Wang X, et al. circRNA CDR1as Promotes Pulmonary Artery Smooth Muscle Cell Calcification by Upregulating CAMK2D and CNN3 via Sponging miR-7-5p. Mol Ther Nucleic Acids 2020;22:530-41. [Crossref] [PubMed]
- Guo J, Zhang L, Lian L, et al. CircATP2B4 promotes hypoxia-induced proliferation and migration of pulmonary arterial smooth muscle cells via the miR-223/ATR axis. Life Sci 2020;262:118420. [Crossref] [PubMed]
- Zhang Y, Chen Y, Yao H, et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ 2019;9:2045894019888416. [Crossref] [PubMed]
- Wang W, Jiang Z, Zhang D, et al. Comparative Transcriptional Analysis of Pulmonary Arterial Hypertension Associated With Three Different Diseases. Front Cell Dev Biol 2021;9:672159. [Crossref] [PubMed]
- Zhang JR, Sun HJ. LncRNAs and circular RNAs as endothelial cell messengers in hypertension: mechanism insights and therapeutic potential. Mol Biol Rep 2020;47:5535-47. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Zhou S, Jiang H, Li M, et al. Circular RNA hsa_circ_0016070 Is Associated with Pulmonary Arterial Hypertension by Promoting PASMC Proliferation. Mol Ther Nucleic Acids 2019;18:275-84. [Crossref] [PubMed]
- Miao R, Gong J, Zhang C, et al. Hsa_circ_0046159 is involved in the development of chronic thromboembolic pulmonary hypertension. J Thromb Thrombolysis 2020;49:386-94. [Crossref] [PubMed]
- Liu R, Zhao W, Wang H, et al. Long Noncoding RNA LINC01207 Promotes Colon Cancer Cell Proliferation and Invasion by Regulating miR-3125/TRIM22 Axis. Biomed Res Int 2020;2020:1216325. [Crossref] [PubMed]
- Chaoyang Y, Qingfeng B, Jinxing F. MiR-216a-5p protects 16HBE cells from H2O2-induced oxidative stress through targeting HMGB1/NF-kB pathway. Biochem Biophys Res Commun 2019;508:416-20. [Crossref] [PubMed]
- He Y, Shi Y, Liu R, et al. PELI3 mediates protumor actions of down-regulated miR-365a-5p in non-small cell lung cancer. Biol Res 2019;52:24. [Crossref] [PubMed]
- Hufbauer M, Maltseva M, Meinrath J, et al. HPV16 increases the number of migratory cancer stem cells and modulates their miRNA expression profile in oropharyngeal cancer. Int J Cancer 2018;143:1426-39. [Crossref] [PubMed]
- Yang H, Huang Y, He J, et al. MiR-486-3p inhibits the proliferation, migration and invasion of retinoblastoma cells by targeting ECM1. Biosci Rep 2020;40:BSR20200392. [Crossref] [PubMed]
- Li C, Zhang Z, Xu Q, et al. Comprehensive Analyses of miRNA-mRNA Network and Potential Drugs in Idiopathic Pulmonary Arterial Hypertension. Biomed Res Int 2020;2020:5156304. [Crossref] [PubMed]
- Wu D, Talbot CC Jr, Liu Q, et al. Identifying microRNAs targeting Wnt/β-catenin pathway in end-stage idiopathic pulmonary arterial hypertension. J Mol Med (Berl) 2016;94:875-85. [Crossref] [PubMed]
- Wei C, Kim IK, Li L, et al. Thymosin Beta 4 protects mice from monocrotaline-induced pulmonary hypertension and right ventricular hypertrophy. PLoS One 2014;9:e110598. [Crossref] [PubMed]
- Chelladurai P, Dabral S, Basineni SR, et al. Isoform-specific characterization of class I histone deacetylases and their therapeutic modulation in pulmonary hypertension. Sci Rep 2020;10:12864. [Crossref] [PubMed]
- Zhang JR, Sun HJ. MiRNAs, lncRNAs, and circular RNAs as mediators in hypertension-related vascular smooth muscle cell dysfunction. Hypertens Res 2021;44:129-46. [Crossref] [PubMed]
- Xiao MS, Ai Y, Wilusz JE. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol 2020;30:226-40. [Crossref] [PubMed]
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-90. [Crossref] [PubMed]


