Lower airway microbiome of children with recurrent wheezing: a clinical cohort study
Introduction
Asthma is a common chronic disease in childhood, with a prevalence of 5–10% (1). Studies have shown that the development of childhood asthma occurs early in life and often manifests as recurrent wheezing (2,3). The syndrome results from a complex interplay between genetic and environmental factors (1). Viral infections have been repeatedly and consistently associated with wheezing episodes, together with factors such as allergy and genetic susceptibility (3).
With the advancement of microbiome detection technologies, the composition of complex microbial communities can be determined by Deoxyribonucleic acid (DNA) sequencing of the conserved bacterial 16S ribosomal Ribonucleic Acid (rRNA) gene, which cannot be detected through traditional bacterial culturing. Hence, several studies have focused on airway microorganisms and demonstrated that a regular lung microbiome exists without any inflammatory or infectious background (4-6). In recent years, comparisons between healthy and wheezing infants have revealed significant differences in several bacterial phylotypes in the upper airways (7). Wheezing episodes are significantly associated with the presence of Haemophilus influenzae and Moraxella catarrhalis in sputum samples, but are independent of viral infections (8). The composition of the microbiota in the upper airways differs from that in the lower airways (9-11), and there is still a paucity of data on the microbial environment of the lower airway of children with recurrent wheezing.
Thus, in this study, we sought to investigate the bacterial flora profile in the lower airways of different patients in Chengdu, Southwest of China, analyse their diversity at a taxonomic level, and investigate the dominant interactions. We tested the hypothesis that the characteristics of the airway microbiome are different in patients with wheezing. Considering that the relationship between the microbiome and the human host has not yet been clarified (12), identifying the risk factors of early recurrent wheezing is of hugely important to the prevention and treatment of childhood asthma. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-165/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Chengdu Women’s and Children’s Central Hospital [No. 2016(22)]. Informed consent was taken from all the patients’ guardians between January 2019 and December 2020.
The study population consisted of children aged 1–3 years with different wheezing symptoms (wheezing frequency ≥ two times or persistent wheezing ≥1 month) who had not used inhaled corticosteroids or antibiotics within 1 week prior to the study, and had been diagnosed by a physician specializing in paediatric pulmonology. The control group comprised children admitted to Chengdu Women and Children’s Central Hospital for the surgical removal of an inhaled bronchial foreign body within 24 hours. Children with known underlying chronic conditions, such as congenital heart diseases, cystic fibrosis, immune deficiency, bronchopulmonary dysplasia, and neuromuscular disorders, were excluded. For children without any apparent explanation for persistent wheezing and without abnormalities on X-ray or computed tomography (CT), a workup including fiberoptic bronchoscopic bronchoalveolar lavage (BAL) was conducted. For these cases, BAL was always performed under stable conditions, avoiding episodes of acute infection or exacerbation of respiratory symptoms (such as fever, polypnea, or dyspnoea)
Alveolar lavage was performed with a fiberoptic bronchoscope (Japan Olympus BF-XP 260 Type F) through a laryngeal mask airway (LMA). The bronchoscope was inserted into one of the segments of the lower lung lobe, and was tightly confined in the sub-segment bronchial opening to prevent the atmospheric mixture of tract secretions and the extravasation of lavage fluid. The BAL samples were snap-frozen on dry ice, shipped to the laboratory, and stored at −80 °C for >6 months prior to DNA extraction.
Twelve kinds of allergen skin prick liquids (including dust mites, house dust mites, cockroaches, cat hair, dog hair, mug wort, yeast, egg white, egg yolk, milk, peanuts, and sea shrimp) obtained from Aroger (Germany) were used for the skin prick test (also from Aroger (Germany). Patients were allergic to a specific substance if the wheal induced by that allergen had a diameter that was at least 25% that of the histamine-induced wheal.
16S rRNA sequencing
The total genomic DNA from the samples was extracted using the CTAB/SDS (Cetyltrimethylammonium Bromide /Sodium dodecyl sulfate) method. The 16S rRNA community profiles were characterised by sequencing (Illumina NovaSeq 6000 system, Novogene Co., Ltd., Beijing, China) of the V3 and V4 regions (insert size 300 bp, read length 250 bp). The sequencing data were filtered to allow each sample to have valid data for 87,349 tags. The effective tags of all the samples were clustered, and sequences with similarities above 97% were considered to be one operational taxonomic unit (OTU). Sequence identities and chimeras were removed using UPARSE. For each representative sequence, the GreenGene database was used to annotate the taxonomic information (13,14).
Bacterial diversity analysis
The DNA was extracted from 5 mL of BALF using the SDS method. The DNA was diluted to 1 ng/µL using sterile water. The 16S ribosomal RNA (rRNA) gene was analysed for bacterial diversity using Illumina Novaseq 6000Alpha diversity rarefactions were calculated by observed species diversity estimates and compared using a two sample t-test. Individual samples were raried based on alpha-diversity estimates, to ensure even sequencing depth for diversity and relative abundance measurements. Beta diversity metrics were calculated using Unweighted UniFrac, and Wilcoxon test for visualization of bacterial communities.
Statistical analysis
All data were analysed using a one-way analysis of variance (ANOVA), followed by a Newman-Keuls multiple comparison test for independent experiments. The results were expressed as the mean ± standard deviation (SD). Statistical differences between the experimental and control groups were examined, and statistical significance was set at P<0.05. SPSS version 20.0 (IBM, Armonk, NY, USA) was used for statistical analysis. Curve-fitting was performed using GraphPad Prism 5 (GraphPad, San Diego, CA, USA).
Results
Patient characteristics
Forty-five hospitalised children with different wheezing symptoms requiring fibreoptic bronchoscopy were initially enrolled. Of these, 13 patients with trachea bronchomalacia were excluded. The final study population included 32 participants (group A). The control group included 23 children who were admitted to the hospital for fibreoptic bronchoscopy within 24 hours following the inhalation of a foreign body (group B). The clinical data showed no significant differences in age, sex, bronchoalveolar lavage neutrophils and eosinophil counts, blood neutrophils and eosinophil counts, and respiratory rate (rate/min) between groups A and B (P>0.05, Table 1). There were marked differences in the gestational duration, mode of delivery, and allergy status between the two groups.
Table 1
| Parameters | Group A | Group B | P value |
|---|---|---|---|
| Number | 32 | 23 | |
| Age (month) | 15.96±5.8 | 18.52±6.14 | 0.124* |
| Sex (male/female) (number) | 19/13 | 14/9 | 0.804# |
| Gestational weeks (≥37/<37 weeks) (number) | 22/10 | 21/2 | 0.0458# |
| Mode of delivery (Natural childbirth/Caesarean) (number) | 14/18 | 17/6 | 0.0261# |
| Allergy status (+/−) (number) | 20/12 | 1/22 | 0.0000# |
| Bronchoalveolar lavage eosinophils (%) | 0.625±1.707 | 0.294±1.212 | 0.4296* |
| Bronchoalveolar lavage neutrophils (%) | 35.5±31.9 | 24.05±20.84 | 0.1194* |
| Blood eosinophils (%) | 2.05±1.7 | 1.84±1.21 | 0.6147* |
| Neutrophil count ×106/g | 9.53±3.19 | 9.37±2.5 | 0.8421* |
| Respiratory rate (rate/min) | 32.00±7.03 | 31±4.03 | 0.5427* |
Data are presented as n, n (%), mean ± SD, and median (quartile 1, quartile 3) unless otherwise indicated. *, t-test; #, Chi-square test.
Overview of species annotation
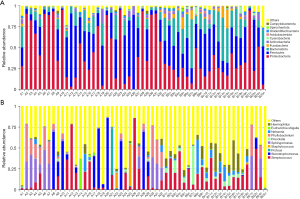
The Illumina MiSeq sequencing platform was used to sequence the V4 hypervariable region of the 16S rRNA for the microbial community in the BAL sample. The sequencing data were filtered to allow each sample to have valid data for 87,349 tags. The effective tags of all samples were clustered, and sequences with similarities above 97% were considered to be one OTU. On average, 6,644 OTUs were detected in every sample. Based on the annotation results, 2,608 (39.25%) OTUs were annotated at the genus level. The dominant phyla were Proteobacteria, Firmicutes, and Bacteroidetes. The dominant classes were Bacilli, Gammaproteobacteria, and Alphaproteobacteria. The dominant orders were Lactobacillales, Xanthomonadales, and Enterobacterales. The dominant families were Streptococcaceae, Xanthomonadaceae, and Morganellaceae. The dominant genera were Streptococcus, Stenotrophomonas, and Proteus. Figure 1 shows a histogram of the relative abundance of species at the phylum and genus levels.
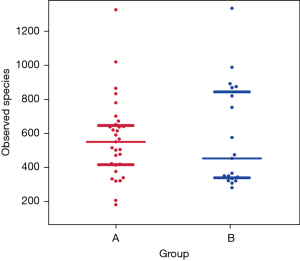
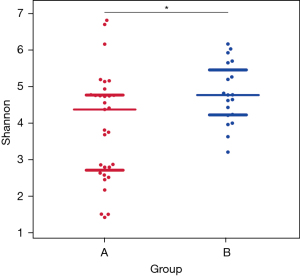
α diversity of the lower airway microbial community
Figure 2 shows the scattered distribution and richness of all species in the two patient groups. A Wilcoxon test revealed that the number of species did not differ significantly between groups A and B (P=0.8574). Figure 3 shows a comparison chart for the Shannon index (diversity index), which reflects differences in the species diversity and their distribution among the different samples. There were marked differences between groups A and B (P=0.0095).
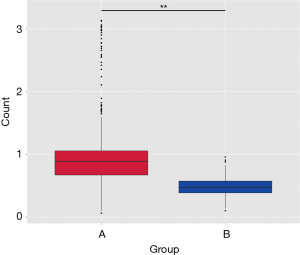
β diversity of the lower airway microbial community
Principal component analysis, Unweighted UniFrac, and Wilcoxon test were conducted to eliminate the individual differences in both groups. Figure 4 displays a box plot based on the diversity of the Unweighted UniFrac Beta, which showed significant differences in β diversity between groups A and B (P=0.00001).
Composition of the lower airway microbial community
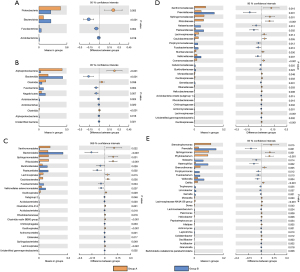
β diversity refers to the comparison of the composition of microbial communities, and the evaluation of the differences between microbial communities. Regarding the species abundance at different levels, different species can be detected using conventional tests. At the phylum level, Proteobacteria accounted for 46.7% of the microbiome of group A and an average of 25.5% of that of group B. We found that the P value for the difference in the species composition of the lung microbiome between the two groups was 0.003. The species with notable differences related to their phyla were Proteobacteria (P=0.003), Bacteroidota (P<0.001), Fusobacteriota (P=0.004), and Acidobacteriota (P=0.019). The species with significant differences related to their genus were Stenotrophomonas (P=0.015), Prevotella (P=0.001), Sphingomonas (P=0.001), Phyllobacterium (P<0.001), Neisseria (P=0.014), and Haemophilus (P=0.002) (Figure 5).
Discussion
Approximately 50% of children experience wheezing within the first year of life, and wheezing phenotypes have been used to identify the characteristics and risk factors associated with wheezing in children (15). Some children suffer from wheezing until late childhood, while others have continuous wheezing throughout adolescence and adulthood. However, the relationship between the risk factors and subsequent development of asthma during later childhood and adult life remains unclear. The important research question being addressed in the current study is the possible correlation between specific patterns of microbial colonization in the respiratory tract and recurrent wheezing/asthma (16). Studies on 16S rRNA and shotgun metagenomics have revealed that a resilient microbiome is present in the upper and lower respiratory tracts, and plays a key role in respiratory diseases (6,7,17). Therefore, the lungs are not considered sterile, and lung diseases have not been exclusively studied in the context of bacterial pathogenesis (17). The density of bacterial communities reduces from the upper respiratory tract to the lower respiratory tract (18,19).
Our study analysed bacterial communities using BAL without the influence of the upper respiratory tract, which only few studies have conducted in the past. We performed this study because the bacterial topographies of the airways of healthy and wheezing children need to be compared. This study demonstrated that the three dominant phyla routinely found in the lower airway of both healthy and wheezing children were Proteobacteria, Firmicutes, and Bacteroides, which are also the dominant phyla of the intestinal microbiome species (20). Consistent with the previous report on healthy airways (19), mature gastrointestinal and respiratory tracts have different environments and functions; however, they have the same embryonic origin and similar structures, and thus, similar changes may also occur. Several chronic diseases of the respiratory tract and intestines have been studied. Although, lung axis disorders are related, the source of lung microecology has not been established, as species may reach the lungs through respiration and oral aspiration.
This study also confirmed that the intestinal lung microecology is consistent. There were differences in the bacterial flora of the two groups based on class, order, family, and genus. Therefore, community richness did not differ between groups A and B (P>0.05). However, there was a marked difference (P<0.05) in the community diversity; α and β diversity were lower in group A than in group B. A previous study also reported that there is a lower α diversity in the flora of untreated asthma patients compared to healthy controls (21). This suggests that flora composition disorders have an impact on the development and progression of recurrent wheezing in children.
Changes in various factors associated with the life course of the host can cause changes in the body’s microecology. This study showed that there were no differences in sex, age, routine blood examination results, and airway inflammatory cells between children with recurrent wheezing and the control group. However, the rate of premature birth in children with recurrent wheezing was higher than those of group B. Several microbial groups have been observed in the placenta, lungs, and intestines in foetuses at 10–18 weeks of gestation through 16S rRNA sequencing, and the foetal lung microbiota matures as the gestational age increases (17,22), suggesting that the airway microecology of newborns at different gestational weeks may differ. A study on the airway microecology of premature infants suggested that the early airway microbiome may prime the developing pulmonary immune system, and the dysbiosis of the microbiome in the airway may set the stage for subsequent lung disease (23). The innate and adaptive immunity of children born via caesarean section is lower than that of normal children, mainly due to the decrease in Th1 (Helper T cell 1) cytokines (24). Differences in immunity may occur due to variations in the microecological compositions of the host (25). We hypothesised that dysbiosis of the lower airway microecology might be a risk factor for recurrent wheezing and asthma in premature infants. Infants born via caesarean section are more likely to have recurrent wheezing than normal infants, which is also related to the variations in lower airway microecology. However, this result should be confirmed by further clinical research.
We also observed enrichment of organisms belonging to the phylum Proteobacteria in patients with recurrent wheezing, including organisms belonging to the Alphaproteobacteria and Bacteroidia classes. Proteobacteria have also been shown to be present in higher proportions in the asthmatic airways of adults (26,27); this bacterial phylum includes common gram-negative respiratory pathogens, such as Haemophilus and Klebsiella pneumonia, which are consistently associated with an increased risk of developing asthma (28). Patients with recurrent wheezing showed a decrease in the number of species belonging to the Bacteroidia (P<0.01), Fusobacteria (P=0.04), and Actinobacteria (P=0.019) phyla. It has been reported that Actinobacteria in the bronchial airway may be protective against bronchial inflammation (29). Our findings also support this notion. Furthermore, a higher abundance of Fusobacteria was previously detected among the bronchial bacterial microbiota in adult asthma patients (30), but a lower abundance of Fusobacteria was detected in the recurrent wheezing group in this study.
Given the anatomical differences in microbiota composition between the upper and lower airway compartments, it is possible to identify several differences between young children and adults (31,32). Therefore the discovery of beneficial airway bacteria may facilitate better treatment of recurrent wheezing in children and aid in the prevention of asthma progression in older children and adults, for example, supplementing with beneficial bacteria. We also found that children in the recurrent wheezing group had a higher incidence of allergic symptoms than those in the control group, which is consistent with previous research (33). This may confirm the similarity between atopic patients with wheezing and asthmatic patients (34), which is beneficial to accurately diagnose wheezing. Toll-like receptor 5 (TLR5) promotes allergic asthma development related to the strong allergic airway response stimulated by bacterial flagellin (35). Our study suggested that an imbalanced bacterial microecology may be one of the reasons for recurrent wheezing. The absence of specific bacterial species early in life could influence appropriate regulatory mechanisms later in life and subsequently shift the immunological balance towards allergy instead of tolerance. Patients in group A were more likely to have asthma and airway hyperresponsiveness than those in group B. Microbiota dysbiosis in the human airway can cause allergy and airway hyperresponsiveness (35). However, the pathogenic mechanism is not fully understood (36).
There were several limitations in this study that could affect the interpretation of our findings. Firstly, both premature birth and caesarean section have been shown to change the microbiome, but since our research sample size was slightly insufficient, further subgroup analysis was of little significance. In future, the sample size needs to be further expanded to confirm the current data. Furthermore, healthy children cannot be used as a control group for ethical reasons. Other limitations of this study include environmental factors such as breastfeeding, number of siblings, attendance of day care, and presence of pets at home, all of which can affect the respiratory microbiome.
Clinical wheezing phenotypes can overlap and change over time. Maintaining a balanced bacterial microecology may be conducive to the prevention of asthma. However, longitudinal studies are required to determine the true nature of the relationship between the microbiome and recurrent wheezing and asthma.
Acknowledgments
Funding: Financial support was provided by the Health Commission of Sichuan Province, China (No. 16PJ067).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-165/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-165/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-165/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of Chengdu Women’s and Children’s Central Hospital [No.2016(
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang K, Yang T, Xu J, et al. Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet 2019;394:407-18. [Crossref] [PubMed]
- Bisgaard H, Szefler S. Prevalence of asthma-like symptoms in young children. Pediatr Pulmonol 2007;42:723-8. [Crossref] [PubMed]
- Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol 2017;140:895-906. [Crossref] [PubMed]
- Mitchell AB, Glanville AR. Introduction to Techniques and Methodologies for Characterizing the Human Respiratory Virome. Methods Mol Biol 2018;1838:111-23. [Crossref] [PubMed]
- Whiteside SA, McGinniss JE, Collman RG. The lung microbiome: progress and promise. J Clin Invest 2021;131:150473. [Crossref] [PubMed]
- Yagi K, Huffnagle GB, Lukacs NW, et al. The Lung Microbiome during Health and Disease. Int J Mol Sci 2021;22:10872. [Crossref] [PubMed]
- Robinson PFM, Pattaroni C, Cook J, et al. Lower airway microbiota associates with inflammatory phenotype in severe preschool wheeze. J Allergy Clin Immunol 2019;143:1607-1610.e3. [Crossref] [PubMed]
- Bisgaard H, Hermansen MN, Bønnelykke K, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ 2010;341:c4978. [Crossref] [PubMed]
- Pfeiffer S, Herzmann C, Gaede KI, et al. Different responses of the oral, nasal and lung microbiomes to cigarette smoke. Thorax 2022;77:191-5. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc 2015;12:821-30. [Crossref] [PubMed]
- Kloepfer KM, Deschamp AR, Ross SE, et al. In children, the microbiota of the nasopharynx and bronchoalveolar lavage fluid are both similar and different. Pediatr Pulmonol 2018;53:475-82. [Crossref] [PubMed]
- Abdel-Aziz MI, Vijverberg SJH, Neerincx AH, et al. The crosstalk between microbiome and asthma: Exploring associations and challenges. Clin Exp Allergy 2019;49:1067-86. [Crossref] [PubMed]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069-72. [Crossref] [PubMed]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. [Crossref] [PubMed]
- Spycher BD, Silverman M, Kuehni CE. Phenotypes of childhood asthma: are they real? Clin Exp Allergy 2010;40:1130-41. [Crossref] [PubMed]
- Dick S, Turner S. The Airway Microbiome and Childhood Asthma - What Is the Link? Acta Med Acad 2020;49:156-63. [PubMed]
- Dickson RP, Huffnagle GB. The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLoS Pathog 2015;11:e1004923. [Crossref] [PubMed]
- Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 2017;15:259-70. [Crossref] [PubMed]
- Goleva E, Jackson LP, Harris JK, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med 2013;188:1193-201. [Crossref] [PubMed]
- Venkataraman A, Bassis CM, Beck JM, et al. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 2015;6:02284-14. [Crossref] [PubMed]
- Huang C, Yu Y, Du W, et al. Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin Transl Allergy 2020;10:42. [Crossref] [PubMed]
- Al Alam D, Danopoulos S, Grubbs B, et al. Human Fetal Lungs Harbor a Microbiome Signature. Am J Respir Crit Care Med 2020;201:1002-6. [Crossref] [PubMed]
- Lal CV, Travers C, Aghai ZH, et al. The Airway Microbiome at Birth. Sci Rep 2016;6:31023. [Crossref] [PubMed]
- Jakobsson HE, Abrahamsson TR, Jenmalm MC, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014;63:559-66. [Crossref] [PubMed]
- Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun 2014;82:4596-606. [Crossref] [PubMed]
- Alamri A. Diversity of Microbial Signatures in Asthmatic Airways. Int J Gen Med 2021;14:1367-78. [Crossref] [PubMed]
- Marri PR, Stern DA, Wright AL, et al. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol 2013;131:346-52.e523. [Crossref] [PubMed]
- Kemter AM, Nagler CR. Influences on allergic mechanisms through gut, lung, and skin microbiome exposures. J Clin Invest 2019;129:1483-92. [Crossref] [PubMed]
- Chun Y, Do A, Grishina G, et al. Integrative study of the upper and lower airway microbiome and transcriptome in asthma. JCI Insight 2020;5:133707. [Crossref] [PubMed]
- Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol 2017;140:63-75. [Crossref] [PubMed]
- Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 2011;127:372-381.e3813. [Crossref] [PubMed]
- Pang Z, Wang G, Gibson P, et al. Airway Microbiome in Different Inflammatory Phenotypes of Asthma: A Cross-Sectional Study in Northeast China. Int J Med Sci 2019;16:477-85. [Crossref] [PubMed]
- Zhang Lei, Ao Ait, Xie Cheng, et al. Analysis of structural changes of lower airway microbiota in Young Children with recurrent wheeze. The Journal of Practical Medicine 2020;36:180-4.
- Aguilera AC, Dagher IA, Kloepfer KM. Role of the Microbiome in Allergic Disease Development. Curr Allergy Asthma Rep 2020;20:44. [Crossref] [PubMed]
- Wilson RH, Maruoka S, Whitehead GS, et al. The Toll-like receptor 5 ligand flagellin promotes asthma by priming allergic responses to indoor allergens. Nat Med 2012;18:1705-10. [Crossref] [PubMed]
- Chen X, Qiu C. Respiratory tract mucous membrane microecology and asthma. Ann Transl Med 2019;7:495. [Crossref] [PubMed]