
Screening of differentially expressed genes in children with cerebral palsy and the construction of a network of the effective components of traditional Chinese medicine
Introduction
Cerebral palsy is the most common disabling disease among children, and includes motor and postural development disorders (1). Motor dysfunction is the core symptom of cerebral palsy (1). An epidemiological study shown that the overall prevalence of cerebral palsy is between 15–35% (2). Preterm birth, maternal infection during pregnancy, low body weight, respiratory distress, and amniotic fluid contamination are high-risk factors for neonatal cerebral palsy (3). Cerebral palsy places enormous burdens (both mental and economic) on families and societies (4). The quality of life of children with cerebral palsy is generally low. To date, the pathogenesis of pediatric cerebral palsy has not yet been completely clarified. It is widely believed that cerebral palsy is related to the joint action of multiple factors in the perinatal period, and hypoxia may be the critical link (5).
Traditional Chinese medicine (TCM) has a good effect in treating infantile cerebral palsy. Angelica sinensis, Shenjincao, Achyranthes bidentata, and other drugs promote blood circulation and menstruation (6-8). However, pharmacological mechanism of TCM is not clear. The rise of network pharmacology and high-throughput omics data analysis technology have created conditions for explaining the occurrence and development of diseases from the perspective of system biology and biological network balance, understanding interactions between drugs and the body with the overall aim of improving or restoring biological network balance, and guiding the discovery of new medicines. Unfortunately, the research on the treatment of cerebral palsy with traditional Chinese medicine is limited to a certain drug or a certain target, and there is no macroscopic screening of key gene sets or construction of drug-gene interaction networks.
We screened differentially expressed gene sets in cerebral palsy blood samples based on high-throughput sequencing data. We obtained the active ingredients of TCM for the treatment of cerebral palsy and the gene set of active ingredients in the TCM pharmacology network database. We de-intersected the two gene sets and constructed an interaction network between TCM active ingredients and potential therapeutic target genes. This may provide clues for elucidating the pharmacological mechanism of TCM in the treatment of cerebral palsy and discovering potential therapeutic targets. We present the following article in accordance with the STREGA reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-171/rc).
Methods
Data download
In this study, a whole transcriptome sequencing data set (GSE183021) of the blood samples of children with cerebral palsy was downloaded from the Gene Expression Omnibus (GEO) database. The data set included blood samples from 5 children with cerebral palsy and 5 control subjects. Batch correction and normalization were performed on the data set, and the ribonucleic acid expression profile was summarized, logarithmically transformed, and combined into a matrix. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential gene screening
Fold change (FC) was calculated as follows: the gene expression of blood samples from patients with cerebral palsy/the gene expression of blood samples from control subjects. The differential genes were screened using the rank-sum test. The differentially expressed genes (DEGs) were screened using the following criteria: a |log2FC| >1, and a false discovery rate (FDR) <0.05.
Gene Ontology (GO) enrichment analysis
A GO functional enrichment analysis was carried out for the differential genes, using David data, and an FDR <0.05 was used as the screening condition. The results of the enrichment analysis were visualized using R package (ggplot).
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
The differential genes were analyzed by the KEGG. The KEGG pathway enrichment analysis was carried out using KEGG data, and an FDR <0.05 was used as the screening condition. The results of the enrichment analysis were visualized using R package (ggplot).
Construction of effective components and target gene network of TCM
The effective components and target genes of TCM were identified from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP). The target genes of TCM combined with the cerebral palsy DEGs from the blood samples were collected, and all the data were imported into Cytoscape 3.61. The effective components and a target gene network of TCM were constructed using the software.
Statistical analysis
All the statistical data in this study were analyzed by R (V3.5.1) and related R package. An FDR <0.05 was considered statistically significant.
Results
Screening of DEGs in cerebral palsy
Taking a | log2FC| ≥1 and an FDR <0.05 as the screening criteria, 399 DEGs were screened from the blood samples of 5 children with cerebral palsy and 5 control subjects. A total of 209 genes were downregulated in the blood samples of children with cerebral palsy, and 190 genes were upregulated. The heat map of the DEGs between the cerebral palsy and control subjects is shown in Figure 1. The top 20 genes with the smallest FDRs are shown in Table 1.
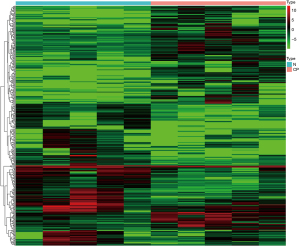
Table 1
| Gene | Cerebral palsy | Control | Log2FC | FDR |
|---|---|---|---|---|
| CXCL8 | 3.23 | 0.03 | 6.67 | 0.001 |
| UNC45B | 2.16 | 0.02 | –6.60 | 0.001 |
| ARC | 17.87 | 0.23 | –6.28 | 0.001 |
| TNF | 23.41 | 0.37 | 5.99 | 0.001 |
| TCEAL6 | 1.37 | 0.02 | 5.94 | 0.001 |
| IL1B | 45.14 | 3.08 | –5.56 | 0.002 |
| NOS2 | 3.02 | 0.10 | 7.96 | 0.002 |
| PEBP4 | 1.62 | 0.06 | –4.88 | 0.002 |
| IL4 | 0.69 | 0.02 | 6.83 | 0.002 |
| RSPO2 | 4.51 | 0.16 | –4.78 | 0.002 |
| BAX | 1.57 | 0.06 | 4.74 | 0.003 |
| VEGFD | 6.91 | 0.28 | –4.60 | 0.003 |
| F7 | 0.56 | 0.02 | 4.49 | 0.003 |
| SFRP5 | 6.51 | 0.29 | –4.47 | 0.003 |
| ADRB3 | 1.26 | 0.06 | 5.41 | 0.004 |
| CMTM5 | 0.80 | 0.04 | –6.38 | 0.004 |
| CCL5 | 0.66 | 0.03 | 2.38 | 0.004 |
| CDK5R2 | 0.87 | 0.05 | –3.28 | 0.004 |
| PCOLCE2 | 4.78 | 0.25 | –4.24 | 0.004 |
| MAPK8 | 0.42 | 0.02 | –4.23 | 0.004 |
DEGs, differentially expressed genes; FDR, false discovery rate; FC, fold change.
GO enrichment analysis of the DEGs in cerebral palsy
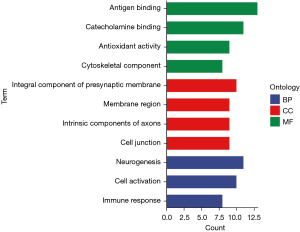
A GO enrichment analysis was performed on the top 20 DEGs in the cerebral palsy samples with the smallest FDRs. The cerebral palsy DEGs were mainly related to immune response, cell activation, and neurogenesis. In terms of the biological processes, the DEGs were primarily associated with the inherent components of axons, membrane regions, presynaptic membrane components, cytoskeleton components, and cell connections. In terms of the cellular components, the DEGs were mainly related to antigen binding, catecholamine binding, antioxidant activity, and the cytoskeleton components. The results are shown in Figure 2.
KEGG enrichment analysis of DEGs in cerebral palsy
A KEGG enrichment analysis was performed on the top 20 cerebral palsy DEGs with the smallest FDRs. Taking an FDR <0.05 as the screening criteria, 11 significantly enriched pathways were ultimately identified. The signaling pathways involved in the immune system and inflammatory response included Th1 cell differentiation, the toll-like receptor signaling pathway, the Nucleotide-binding and oligomerization domain (NOD)-like receptor signaling pathway, Th1 and Th2 cell differentiation, natural-killer cell-mediated cytotoxicity, antigen processing and presentation, and cytokine receptor interaction. The signaling pathways involved in the nervous system included the phosphatidylinositol 3-kinase (PI3K)-Akt signal pathway, and the hypoxia-inducible factor-1 (HIF-1) signaling pathway. Other pathways included the cell cycle, and the cytoplasmic deoxyribonucleic acid (DNA) sensing pathway. The KEGG enrichment analysis showed that the cerebral palsy DEGs were mainly related to the immune and inflammatory response signaling pathways and the nervous system signaling pathways. The results are shown in Figure 3.

Prediction of effective components of TCM
A database analysis of the effective components of TCM revealed that 3 effective ingredients (i.e., Angelica sinensis, Shenjincao, and Achyranthes bidentata) were mainly used to treat infantile cerebral palsy. The results are shown in Table 2.
Table 2
| Drug | ID | Name |
|---|---|---|
| Angelica sinensis | MOL004214 | (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one |
| MOL003564 | Mandenol | |
| MOL003541 | Ethyl oleate (NF) | |
| MOL001833 | Beta-sitosterol | |
| MOL002455 | Stigmasterol | |
| MOL002186 | Quercetin | |
| MOL000643 | 1-Monolinolein | |
| MOL000586 | ((1S)-3-((E)-but-2-enyl)-2-methyl-4-oxo-1-cyclopent | |
| MOL001674 | Sexangularetin | |
| MOL004932 | Beta-sitosterol | |
| MOL000936 | Sitosterol | |
| MOL000421 | Berberine | |
| Shenjincao | MOL002471 | Berberrubine |
| MOL008611 | Epiberberine | |
| MOL006531 | (R)-Canadine | |
| MOL000231 | Berlambine | |
| MOL000912 | Corchoroside A_qt | |
| Achyranthes bidentata | MOL001340 | Magnograndiolide |
| MOL000784 | Palmatine | |
| MOL000936 | Sitosterol | |
| MOL000421 | Berberine | |
| MOL000912 | Corchoroside A_qt |
Intersection of target genes and the differential genes of TCM
The active components of Angelica sinensis, Shenjincao, and Achyranthes bidentata, targeted 158 genes, and 49 genes crossed with the cerebral palsy DEGs (see Figure 4).

TCM and construction of differential gene network
The active components of Angelica sinensis, Shenjincao, and Achyranthes bidentata, and the cerebral palsy DEGs were taken as the nodes to construct the action network (see Figure 5). Interleukin (IL)-1B, IL-6, prostaglandin-endoperoxide synthase 1 (PTGS1), tumor necrosis factor (TNF), estrogen receptor 1 (ESR1), nitric oxide synthase 2 (NOS2), and other genes played an important role with the effective components of TCM.

Discussion
This study analyzed a transcriptome sequencing data set of blood samples of children with cerebral palsy, screened the DEGs in the blood samples of the children with cerebral palsy and control subjects. We have 190 up-regulated genes and 209 down-regulated genes. We believe that there are more down-regulated genes than up-regulated genes, and the sample size may be related to our screening criteria. We conducted an enrichment analysis of those genes. The GO and KEGG enrichment analyses showed that these DEGs were related to the immune response, neurogenesis, inherent components of axons, presynaptic membrane components, cell connection, antioxidant activity, immune response, and nervous system signaling pathways. Our results provide insights into the pathogenesis of DEGs, and confirmed the representativeness of these DEGs, which may play key roles in the pathogenesis of cerebral palsy.
Consistent with our results, previous studies (4,5) have shown that cerebral palsy may be related to natural-killer cell-mediated cytotoxicity, and antigen processing and presentation. This study obtained the relevant information about the TCM components and targets, such as Angelica sinensis, Shenjincao, and Achyranthes bidentata, from the TCMSP. An interaction network was then constructed by combining the DEGs. IL1B, IL6, PTGS1, TNF, toll-like receptor 4 (TLR4), progesterone receptor (PGR), nitric oxide synthase (NOS2), and other genes were shown to play an important role in the effective TCM components. We also found that these target genes were closely related to nervous system development and diseases.
Additionally, we found that IL-6, IL-1B, and TNF are the best protein biomarkers for predicting neurodevelopment injury (9). IL-6 is an important neuroinflammatory factor that mediates brain inflammatory lesions and amyloidosis, and is associated with a variety of brain diseases, including Alzheimer’s disease, and autism (10,11). IL-6 is associated with periventricular leukomalacia and may affect neurodevelopment and increase the risk of cerebral palsy (12). Studies have shown that in children with cerebral palsy, cytokines are dysregulated, and plasma TNF levels are elevated (13,14). IL-1B plays a key role in inflammatory demyelinating polyradiculoneuropathy (15).
A Study also confirmed that the amplification of microsatellites in the IL-1B promoter and the NOS2A promoter may make children more prone to cerebral palsy after hypoxic-ischemic encephalopathy (16). TLR4 belongs to the type I transmembrane protein, which is activated through both MyD88 dependent and non-MyD88 dependent pathways, which activate macrophages and endothelial cells to promote the expression of inflammatory and immunomodulatory factor genes. A clinical study shown that TLR4 polymorphism has a potential neuroprotective effect, which may be related to reducing the risk of cerebral palsy (17).
PTGS1 is considered a target for relieving pain and treating inflammation. In the neuroinflammatory response, the expression of PTGS1 in astrocytes and microglia is significantly increased (18). PTGS1 may be a pathological factor leading to neurodegeneration, mental disease, and epilepsy (18-20).
NOS2 promotes leukocyte migration to inflammatory sites via the endothelium and destroys the blood-brain barrier. The upregulation of NOS2 occurs in various diseases, including autoimmune diseases, cancer, cardiovascular diseases, nervous system diseases, and inflammatory diseases (21-23). Under inflammatory conditions, elevated NOS2 increases the risk of white-matter damage and cerebral palsy in children.
By combining GEO and TCMSP data, we constructed an action network with the active ingredients of Angelica sinensis, Shenjincao, Achyranthes bidentata and cerebral palsy differentially expressed genes as nodes, and screened out IL1B, IL6, PTGS1, TNF, TLR4, PGR, NOS2 and other genes. These genes are all potential therapeutic targets. We believe that functional studies can be carried out based on these genes to verify the interaction between the effective components of TCM and genes, so as to determine therapeutic targets and innovate new drugs.
There are some flaws in this study. The first is the small amount of cerebral palsy blood samples used to analyze differential genes in this study, which needs to be further expanded with large sample size for verification. Second, this study lacks experimental validation in vivo and in vitro to support the results of our analysis. Finally, functional experiments of key genes need to be carried out to clarify the pathogenic mechanism and the mechanism of drug action. In sum, this study used a transcriptome sequencing data set to screen the DEGs of cerebral palsy samples. We constructed an action network with the active components of Angelica sinensis, Shenjincao, Achyranthes bidentata, and cerebral palsy DEGs as nodes by combining with the pharmacological network database of TCM. Our findings can be used to further clarify the pharmacological mechanism of TCM in the treatment of cerebral palsy and identify novel therapeutic targets.
Acknowledgments
Funding: The study was supported by the Natural Science Foundation of Zhejiang Province (No. LQ20H090009), National Center Independent Design Project, Multi Center Clinical Research Project in Zhejiang Province (No. S20A0002) and Zhejiang Province Traditional Chinese Medicine Science and Technology Project (No. 2022ZB182).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-171/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-171/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Vitrikas K, Dalton H, Breish D. Cerebral Palsy: An Overview. Am Fam Physician 2020;101:213-20. [PubMed]
- Green MM, Gaebler-Spira D. Cerebral Palsy. J Pediatr Rehabil Med 2019;12:113-4. [Crossref] [PubMed]
- Morgan C, Fetters L, Adde L, et al. Early Intervention for Children Aged 0 to 2 Years With or at High Risk of Cerebral Palsy: International Clinical Practice Guideline Based on Systematic Reviews. JAMA Pediatr 2021;175:846-58. [Crossref] [PubMed]
- Gaebler-Spira D, Green M. Cerebral Palsy. J Pediatr Rehabil Med 2020;13:105-6. [Crossref] [PubMed]
- Appleton RE, Gupta R. Cerebral palsy: not always what it seems. Arch Dis Child 2019;104:809-14. [Crossref] [PubMed]
- Liao HH, Yen HR, Muo CH, et al. Complementary traditional Chinese medicine use in Children with cerebral palsy: a nationwide retrospective cohort study in Taiwan. BMC Complement Altern Med 2017;17:155. [Crossref] [PubMed]
- Zhang Y, Liu J, Wang J, et al. Traditional Chinese Medicine for treatment of cerebral palsy in children: a systematic review of randomized clinical trials. J Altern Complement Med 2010;16:375-95. [Crossref] [PubMed]
- Wu Y, Zou LP, Han TL, et al. Randomized controlled trial of traditional Chinese medicine (acupuncture and tuina) in cerebral palsy: part 1--any increase in seizure in integrated acupuncture and rehabilitation group versus rehabilitation group? J Altern Complement Med 2008;14:1005-9. [Crossref] [PubMed]
- Nist MD, Pickler RH. An Integrative Review of Cytokine/Chemokine Predictors of Neurodevelopment in Preterm Infants. Biol Res Nurs 2019;21:366-76. [Crossref] [PubMed]
- Flores-Aguilar L, Iulita MF, Orciani C, et al. Cognitive and brain cytokine profile of non-demented individuals with cerebral amyloid-beta deposition. J Neuroinflammation 2021;18:147. [Crossref] [PubMed]
- Thadathil N, Nicklas EH, Mohammed S, et al. Necroptosis increases with age in the brain and contributes to age-related neuroinflammation. Geroscience 2021;43:2345-61. [Crossref] [PubMed]
- Grubač Ž, Šutulović N, Jerotić D, et al. Experimental chronic sleep fragmentation alters seizure susceptibility and brain levels of interleukins 1β and 6. Acta Neurobiol Exp (Wars) 2021;81:96-109. [Crossref] [PubMed]
- Zareen Z, Strickland T, Fallah L, et al. Cytokine dysregulation in children with cerebral palsy. Dev Med Child Neurol 2021;63:407-12. [Crossref] [PubMed]
- Sharova O, Smiyan O, Borén T. Immunological effects of cerebral palsy and rehabilitation exercises in children. Brain Behav Immun Health 2021;18:100365. [Crossref] [PubMed]
- Langer K, Cohen RA, Porges EC, et al. Circulating Cytokines Predict 1H-Proton MRS Cerebral Metabolites in Healthy Older Adults. Front Aging Neurosci 2021;13:690923. [Crossref] [PubMed]
- Torres-Merino S, Moreno-Sandoval HN, Thompson-Bonilla MDR, et al. Association Between rs3833912/rs16944 SNPs and Risk for Cerebral Palsy in Mexican Children. Mol Neurobiol 2019;56:1800-11. [Crossref] [PubMed]
- Djukic M, Gibson CS, Maclennan AH, et al. Genetic susceptibility to viral exposure may increase the risk of cerebral palsy. Aust N Z J Obstet Gynaecol 2009;49:247-53. [Crossref] [PubMed]
- Cai H, Cai B, Sun L, et al. Association between PTGS1 polymorphisms and functional outcomes in Chinese patients with stroke during aspirin therapy: Interaction with smoking. J Neurol Sci 2017;376:211-5. [Crossref] [PubMed]
- Palma-Barqueros V, Bohdan N, Revilla N, et al. PTGS1 gene variations associated with bleeding and platelet dysfunction. Platelets 2021;32:710-6. [Crossref] [PubMed]
- Wang Y, Liu Y, Zhang M, et al. Inhibition of PTGS1 promotes osteogenic differentiation of adipose-derived stem cells by suppressing NF-kB signaling. Stem Cell Res Ther 2019;10:57. [Crossref] [PubMed]
- Arneson-Wissink PC, Doles JD. Disrupted NOS2 metabolism drives myoblast response to wasting-associated cytokines. Exp Cell Res 2021;407:112779. [Crossref] [PubMed]
- Kobayashi S, Homma T, Fujii J. Nitric oxide produced by NOS2 copes with the cytotoxic effects of superoxide in macrophages. Biochem Biophys Rep 2021;26:100942. [Crossref] [PubMed]
- Zhong J, Scholz T, Yau ACY, et al. Mannan-induced Nos2 in macrophages enhances IL-17-driven psoriatic arthritis by innate lymphocytes. Sci Adv 2018;4:eaas9864. [Crossref] [PubMed]


