Site-directed mutagenesis of neonatal convulsions associated KCNQ2 gene and its protein expression
Benign familial neonatal convulsions (BFNC) is a rare idiopathic epilepsy of newborns with autosomal dominant inheritance. It is characterized by frequent unprovoked seizures that typically begin with in the first days of life and spontaneously disappear within several weeks to months (1). Seizures can be partial or manifest as generalized convulsions that occur during wakefulness and sleep. Patients typically have a normal physical exam and long-term neurodevelopment, however, learning disabilities or delayed speech development have been observed in a few individuals (2). The early neonatal onset distinguishes BFNC from two other types of hereditary focal epilepsies of infancy, benign familial neonatal infantile convulsions (BFNIC) (3) and benign familial infantile convulsions (BFIC) (4). In addition to a later age of onset, BFNIC and BFIC are genetically distinct from BFNC (5).
Benign familial neonatal convulsions (BFNC) is an epileptic disorder caused by dominant mutations in the genes KCNQ2 and KCNQ3. In recent years, research on genes that cause epilepsy has made significant progress. Rett and Teubel reported the first family in 1964 and the association of KCNQ potassium channels with epilepsy has become a focus of interest since the report in Science about the close relationship between mutations of the potassium channel gene KCNQ2 and neonatal convulsions in 1998. In 2003, Tang et al. (6) published the first analysis of KCNQ2 mutation in a benign familial neonatal convulsion (BFNC) pedigree in China, reporting the finding of 1931delG, a new mutation in KCNQ2. our previous study (7) reported a novel missense mutation, c.812G>T, in the KCNQ2 gene of a benign familial infantile convulsion (BFIC) pedigree, which causes replacement of glycine by valine at position 271 of the KCNQ2 channel (p.G271V) among involved children. Seventeen patients have been identified in 62 people of 4 generations in this pedigree. As KCNQ2 mutations interfere in the potassium channels through different pathways, diverse phenotypes may be present as a result of different underlying mutants. And phenotypic variability has been recognized including: later onset of seizures, myokymia in isolation or accompanied by seizures, neurological deficit and mental retardation. However, mutated gene function and the impact of mutations on the corresponding phenotypes are thus critical to successful individual treatment (8). One of the ways to investigate the functions of a mutant is to build an eukaryotic expression vector for the mutant gene and the model for the gene expression, which will be subject to drug studies. Up to now, the ion channel expression experiments of KCNQ2/KCNQ3 in Xenopus oocytes and human embryonic kidney (HEK) cells have been well established (9).
In this study, the KCNQ2 mutation c.812G>T was engineered on KCNQ2 cDNAs by sequence overlap extension PCR and HEK293 cells were transfected with the eukaryotic expression vectors carrying that mutation to establish the cell model for research of the related gene functions in neonatal/infant convulsions as well as further drug research.
Materials and methods
Materials
The following materials were used in this study: eukaryotic expression plasmid pcDNA3.0-KCNQ2 containing full-length KCNQ2-cDNA fragments, provided by Pharmacology Research Center of Hebei Medical University; Vector green fluorescent protein pRK5 (pRK5-GFP), HEK293 cells, and E. coli DH5α, preserved in our laboratory; Taq DNA polymerase (TaKaRa Company); T4 DNA ligase, various restriction enzymes and competent bacteria XL1-Blue (Invitrogen products); gel extraction kit and PCR product purification kit (MBI); liposomal plasmid transfection reagent Superfect (Qiagen); DMEM media (Gibco); goat anti-human KCNQ2-N terminal monoclonal antibodies and rabbit anti-goat IgM-FITC (Pierce USA). Synthesis and sequencing of primers were completed by Oak, Beijing. PTC-200DNA Engine (MJ Research, USA) and Bio-Rad Gel Doc 2000 were used in the study.
Methods
Primer design
Two pairs of primers, P1/P2 and P3/P4, were designed based on the normal KCNQ2-cDNA sequence. P1 was introduced into the EcoRI restriction site while P2 and P3 carried mutations at position 812 (G812T) with a 20 bp sequence complementary to each other, and P4 was introduced into the Hind III restriction site.
The two pairs of primers were: P1: CAGAATTCCTGCAGCCACTCATCCCCGCTGAGCCTGAG (38 mer) (containing EcoRI restriction site); P2: TGATCAGGACCCACCAGAGTGCATC (25 mer) (introduced to the position of mutation); P3: ACTCTGGTGGGTCCTGATCACGCTGACCACCATTG (35 mer) (introduced to the position of mutation); and P4: CTTAAGCTTTGTAAAAGGTCACTGCCAG (28 mer) (containing the Hind III restriction site). The primers synthesized by Beijing Aoke Biological Product Company were prepared to 100 pmol/µL with deionized water.
PCR procedures
Three steps in the PCR were followed. First, 37 µL ddH2O, 5 µL 10× Pfu Polymerase Buffer (with Mg2+), 4 µL dNTP, 1 µL of primers P1 and P2 each, 1 µL (2.5 U) Pfu DNA Polymerase and 1 µL (approximately 100 ng) KCNQ2-cDNA template were in turn added into a 50 µL reaction system. The reaction conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 50 s and extension at 72 °C for 1.5 min, with a last extension at 72 °C for 15 min. Primers P3 and P4 were processed in the same reaction system. PCR products of P12 and P34 were tested against 0.8% agarose gel electrophoresis. After recovery with a gel extraction kit, they were subject to the subsequent reactions. TaKaRa LA Taq was used to connect P12 and P34 to P1234 with purified P12 and P34 as the template and P1, P4 as primers. Into a 50 µL reaction system, 11 µL ddH2O, 5 µL GC Buffer (with Mg2+) ×2, 8 µL dNTP, 1 µL of primers P1 and P2 each, 1 µL of recovered P12 and P34 each, and 1 µL TaKaRa LA Taq enzyme were added. The amplification conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 90 s, annealing at 65 °C for 90 s and extension at 72 °C for 3.5 min, with a last extension at 72 °C for 30 min. A specific band P1234 was identified in the products through 1% agarose gel electrophoresis.
Creation of intermediate vector DNA fragments P1234 were connected to pMD19-T
Simple Vector using T/A cloning. As blunt-ended Pfu PCR products, DNA fragments P1234 should be tailed before T/A cloning. After the final cycle of PCR reaction, 1 µL of 100 mmol/L dATP and 1 µL of 2 U/µL Taq DNA Polymerase were added into the reaction system and heated at 72 °C for 30 min. Blue-white screening was conducted following the instructions on the T/A cloning kit (X-gal/IPTG added to the plate in advance). White spots were inoculated into an ampicillin-containing LB medium and incubated at 37 °C overnight. A small amount of plasmids were extracted the next day using alkaline lysis to verify that the target gene DNA fragments had been connected to pMD19-T Simple Vector.
Creation of the eukaryotic expression vector containing point mutations
Verified plasmid pMD19-T-c.812G>T-KCNQ2 that contained the correct point mutations and wild-type pcDNA3.0-KCNQ2 were double digested using restriction enzymes EcoRI and Hind III, respectively. The digestion products were separated on 0.8% agarose gel electrophoresis. Small fragments of pMD19-T-c.812G>T-KCNQ2 and the large portion of pcDNA3.0-KCNQ2 were recovered by the Qiagen extraction kit. Mixed with a molar ratio of 1:4, those recovered products were incubated with T4 DNA ligase at 16 °C overnight to transfect the host bacteria E.coliDH5α, which were then cultured on an ampicillin-containing LB agar plate overnight. Well growing colonies were inoculated into an ampicillin-containing LB medium and cultured in 37 °C shaker overnight. A small amount of plasmids were extracted by alkaline lysis. The extracted pcDNA3.0-c.812G>T-KCNQ2 was subject to 1% agarose gel electrophoresis and ABI 3700 automated DNA sequencing to verify that the introduction of mutations was successful and there was no other randomly generated mutations.
Transfection of HEK293 cells
One day before transfection, the cells were allowed to grow normally in 2 mL serum-containing, antibiotic-free DMEM medium (6 well plate) to reach a density of 90-95% on the day of transfection. The cells were co-transfected with pcDNA3.0-KCNQ2 and pcDNA3.0-GFP using lipofectamine following the kit instructions. The transfection mixture included: (I) 2 µg pcDNA3.0-c.812G>T-KCNQ2, 1 µg GFP; (II) 2 µg wild-type pcDNA3.0-KCNQ2,1 µg GFP; (III) 3 µg GFP was added into the positive control group only; (IV) 3 µg empty pcDNA3.0 vector was added into the negative control group only; and 10 µL plasmids were added into each group. The cells were first cultured in serum-free DMEM for 3 h and transferred to the serum-containing medium. The result of transfection was observed after 48 h and photo taken under the microscope.
Immunofluorescence detection of KCNQ2 protein expression
To rule out the impact of green fluorescent proteins on the expression and localization of KCNQ2 proteins in cells, immunofluorescent chemical approach was employed to further observe the intracellular expression of wild-type and mutant proteins. Forty-eight hours after transfection, polylysine coated coverslips with cell growth were removed from the dish, washed with PBS buffer, fixed with 4% oligomeric formaldehyde for 15 min, and rinsed with PBS suffer containing 0.1 mg/mL saponin. After store with 5% BSA at room temperature for 10 min, goat anti-human KCNQ2-N terminal polyclonal antibody (1:200 dilution) was added and incubated at room temperature for 2 h. The mixture was then washed with PBS for three times, 5 min for each, incubated at 37 °C in dark for 40 min after addition of FITC-conjugated rabit anti-goat IgG (1:200 dilution), and again washed with PBS for three times, 5 min for each. Mounted in glycerol PBS, the section was then observed under fluorescence microscope.
Results
PCR products
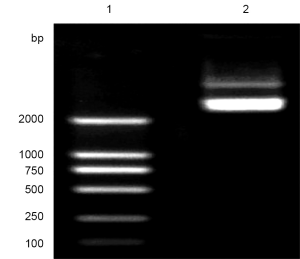
In the first and second round of PCR reactions, fragments P12 (888 bp) and P34 (1912 bp) with sequences complementary to each other were obtained after amplication of primers P1/P2 and P3/P4 using wild-type plasmid pCDNA3.0-KCNQ2 as the template. Both of them had point mutations, and 0.8% agarose gel electrophoresis showed that evident product bands at 888 bp and 1912 bp, i.e. P12 and P34 (Figure 1).

In the second round, 10 ng of equivalent purified P12 and P34 were added as the template for amplification of P1 and P4 to produce P1234 fragment (2,800 bp) with the target mutation. A noticeable product band was shown in the extraction of the purified fragment P1234, which was the product in the overlap PCR (Figure 2).
Creation of the mutant eukaryotic expression vector
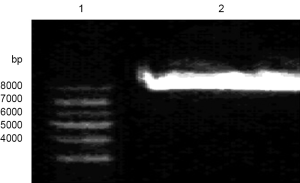
As the products of the third-round PCR were connected into pMD19-T Simple Vector, verified plasmid pMD19-T-c.812G>T-KCNQ2 that contained the correct point mutations was double digested with restriction enzymes EcoRI and Hind III, respectively. The digestion products were separated by agarose gel electrophoresis. As a result, P1234 was reflected at 2,800 bp on the band, which was a mutation-containing DNA fragment. Meanwhile, pcDNA3.0-KCNQ2 was double digested with EcoRI and Hind III, resulting in a target fragment at around 8,000 bp on the band (Figure 3).
Sequencing of point mutation-containing pcDNA3.0-c.812G>T-KCNQ2
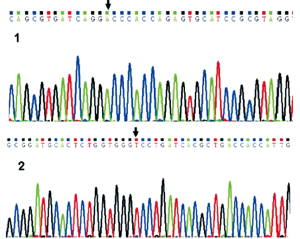
The extracted pcDNA3.0-c.812G>T-KCNQ2 was subject to agarose gel electrophoresis and ABI 3700 automated DNA sequencing, which showed successful point mutation of the target gene on the vector, ie. the nucleotide at position 812 of KCNQ2-cDNA had been changed from G into T without any other sequence changes (Figure 4).
Transfected HEK293 cells under microscope
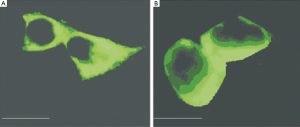
Under the phase contrast microscope, non-transfected HEK293 cells were closely adherent to each other with an irregular, polygonal cell shape. Under the confocal microscope, they did not show green fluorescence, while those co-transfected with pcDNA3.0-KCNQ2 and pRK5-GFP plasmids showed obvious green fluorescence, indicating successful transfection (Figure 5).

Immunofluorescence results
FITC green fluorescent-marked wild-type KCNQ2 and mutant KCNQ2-c.812G>T proteins were both expressed on the cell membrane (Figure 6). The similar immunochemical appearance between wild-type and mutant KCNQ2-c.812G>T potassium channels suggested that the c.812G>T point mutation in KCNQ2 did neither reduce the protein expression nor change protein intracellular location in HEK293 cells.
Discussion
The KCNQ2 protein belongs to the voltage-gated potassium channel family, where members share a highly similar subunit structure -- both containing six hydrophobic transmembrane regions (S1-S6). One voltage sensor is in the conservatively ion selective pore region between S5 and S6, one in the S4 region, and the others in the N- and C-terminals in the cytoplasm. Five subunit genes (KCNQ1-KCNQ5) of the family are known to be able to form an ion channel. KCNQ1 mutation is one of the cause of congenital long QT syndrome (10), and KCNQ4 mutation causes dominant hereditary deafness in children. KCNQ2/KCNQ3 genes, encoding the M-type potassium ion channel in combination, are the cause of BFNC. KCNQ2 mutations are largely associated with reduction in M-type potassium current and consequent increased excitability and explosive discharge of nerve cells (11).
A number of these mutations have been found to be related with epilepsy, most being deletions, insertions or splice site mutations (12). While most of them can result in reading frame shift and a premature stop codon, only a few missense mutations have been reported to be located in the pore region or transmembrane regions 4, 5, and 6 (9). In this study, missense mutation c.812G>T of the KCNQ2 gene was found in a Chinese BFIC pedigree of 17 patients, which was so far the third reported KCNQ2 mutation in the pore region. Past studies suggested that the mutation did not affect the effective expression of KCNQ2 on the cell membrane when located in this region, but it reduced the current magnitude by interfering ionic conductance (13). As shown in our current study, mutant c.812G>T and wild-type KCNQ2 proteins were expressed in HEK293 cells through lipofection, and immunofluorescence confirmed located expression of both mutant and wild-type proteins on the membrane, suggesting that c. 812G>T mutation did not affect the expression of proteins in cells or its transport from the cytoplasm to the cell membrane.
Glycine at position 271 of the KCNQ2 potassium channel was replaced by valine (p.G271V) as a result of c.812G>T mutation, which was a highly conserved position in the pore region of the KCNQ family in many animals. As reported by Charlier et al. (14), p.G271V was an amino acid change in the same position as that in KCNQ3 mutation (p.G310V) of a BFNC pedigree where glycine was also replaced by valine. In the study conducted by Charlier et al. (14) for in vitro expression of wild-type KCNQ3, KCNQ3-p.G310V and KCNQ2 (at the ratio of 1:1:2), a 20-30% decrease in the current amplitude was found when compared with the co-expression of wild-type KCNQ3 and KCNQ2 (1:1). One KCNQ3 mutation, p.W309R, was found to be adjacent to p.G310V in a Japanese pedigree, where zero potassium current was detected from in vitro co-expression of p.W309R and wild-type KCNQ2. The researchers speculated that p.W309R closed the potassium channels and led to potassium ion conduction defects (15). In patients with congenital long QT syndrome, glycine replacement was found at the same position of KCNQ1 (p.G306R) (16). When expressed in vitro, p.G306R was associated with significant reduction in the potassium current, which could not be salvaged by the specific activator R-L3. The researchers believed that the p.G306R mutation might have destroyed the key site for activation of the channels (17).
KCNQ2 mutations were first considered to be related to BFNC with a benign course. However, the mutations were later found in children with drug-resistant epileptic encephalopathy and seizures complicated with mental retardation (18,19). Wang et al. (20) located the genetic cause of a large BFIC pedigree onto chromosome region 20q13.3, where the KCNQ2 gene situated. Ishii et al. (21) found KCNQ2 gene mutations in children with non-benign familial neonatal convulsions; and Neubauer et al. (22) also found multiple KCNQ2/KCNQ3 gene mutations in patients with exercise-induced epilepsy and sporadic idiopathic generalized epilepsy. These findings demonstrated that KCNQ2 mutations were not limited to the typical BFNC pedigree. Therefore, a combination of the genotype, phenotype, and the structure, function and regulation mechanism of mutant proteins should be taken into account.
Currently, KCNQ potassium channels have become new targets for antiepileptic drugs. For example, the M-channel opener, retigabine, was proved effective in animal models of epilepsy. Its Phase I and Phase IIa clinical trials were completed, and a Europe-US multi-center Phase IIb clinical trial was in progress (23). Fluoropyridine was an analgesic agent developed by German scientists. Recent studies found that it could serve as an M-channel opener as retigabine, which could significantly enhance the KCNQ2/KCNQ3 channel current response in HEK293 cells transfected with KCNQ2/KCNQ3 genes. Those two drugs might be useful for the treatment of refractory epilepsy resistant to currently available options.
In summary, a mutant protein expression model was created in this study by engineering c.812G>T on KCNQ2 cDNAs via sequence overlap extension PCR and transfecting HEK293 cells with the eukaryotic expression vectors carrying that mutation. The next step would be focused on the functional differences between mutant and wild-type KCNQ2 genes in cells for better understanding of their roles in pathogenesis of convulsions and providing information for appropriate drug intervention.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ronen GM, Rosales TO, Connolly M, et al. Seizure characteristics in chromosome 20 benign familial neonatal convulsions. Neurology 1993;43:1355-60. [PubMed]
- Lerche H, Weber YG, Jurkat-Rott K, et al. Ion channel defects in idiopathic epilepsies. Curr Pharm Des 2005;11:2737-52. [PubMed]
- Borgatti R, Zucca C, Cavallini A, et al. A novel mutation in KCNQ2 associated with BFNC, drug resistant epilepsy, and mental retardation. Neurology 2004;63:57-65. [PubMed]
- Vigevano F, Fusco L, Di Capua M, et al. Benign infantile familial convulsions. Eur J Pediatr 1992;151:608-12. [PubMed]
- Hunter J, Maljevic S, Shankar A, et al. Subthreshold changes of voltage-dependent activation of the K(V)7.2 channel in neonatal epilepsy. Neurobiol Dis 2006;24:194-201. [PubMed]
- Tang B, Li H, Xia K, et al. A novel mutation in KCNQ2 gene causes benign familial neonatal convulsions in a Chinese family. J Neurol Sci 2004;221:31-4. [PubMed]
- Zhou X, Ma A, Liu X, et al. Infantile seizures and other epileptic phenotypes in a Chinese family with a missense mutation of KCNQ2. Eur J Pediatr 2006;165:691-5. [PubMed]
- Miceli F, Soldovieri MV, Martire M, et al. Molecular pharmacology and therapeutic potential of neuronal Kv7-modulating drugs. Curr Opin Pharmacol 2008;8:65-74. [PubMed]
- Volkers L, Rook MB, Das JH, et al. Functional analysis of novel KCNQ2 mutations found in patients with Benign Familial Neonatal Convulsions. Neurosci Lett 2009;462:24-9. [PubMed]
- Viadero MT, Rubín E, Amigo T, et al. Three generations of hereditary long-QT syndrome with complete penetrance caused by the p.G316E KCNQ1 mutation. Pediatr Cardiol 2011;32:102-4. [PubMed]
- Otto JF, Yang Y, Frankel WN, et al. A spontaneous mutation involving Kcnq2 (Kv7.2) reduces M-current density and spike frequency adaptation in mouse CA1 neurons. J Neurosci 2006;26:2053-9. [PubMed]
- Heron SE, Cox K, Grinton BE, et al. Deletions or duplications in KCNQ2 can cause benign familial neonatal seizures. J Med Genet 2007;44:791-6. [PubMed]
- Uehara A, Nakamura Y, Shioya T, et al. Altered KCNQ3 potassium channel function caused by the W309R pore-helix mutation found in human epilepsy. J Membr Biol 2008;222:55-63. [PubMed]
- Charlier C, Singh NA, Ryan SG, et al. A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat Genet 1998;18:53-5. [PubMed]
- Sugiura Y, Nakatsu F, Hiroyasu K, et al. Lack of potassium current in W309R mutant KCNQ3 channel causing benign familial neonatal convulsions (BFNC). Epilepsy Res 2009;84:82-5. [PubMed]
- Seebohm G, Scherer CR, Busch AE, et al. Identification of specific pore residues mediating KCNQ1 inactivation. A novel mechanism for long QT syndrome. J Biol Chem 2001;276:13600-5. [PubMed]
- Seebohm G, Pusch M, Chen J, et al. Pharmacological activation of normal and arrhythmia-associated mutant KCNQ1 potassium channels. Circ Res 2003;93:941-7. [PubMed]
- Dedek K, Fusco L, Teloy N, et al. Neonatal convulsions and epileptic encephalopathy in an Italian family with a missense mutation in the fifth transmembrane region of KCNQ2. Epilepsy Res 2003;54:21-7. [PubMed]
- Borgatti R, Zucca C, Cavallini A, et al. A novel mutation in KCNQ2 associated with BFNC, drug resistant epilepsy, and mental retardation. Neurology 2004;63:57-65. [PubMed]
- Wang JQ, Li HM, Yin JG, et al. Linkage mapping of benign familialinfantile convulsions(BFIC) and sequencing analysis of KCNQ2 gene in a BFIC family. Zhong hua Er Ke Za Zhi 2002;40:513-7.
- Ishii A, Fukuma G, Uehara A, et al. A de novo KCNQ2 mutation detected in non-familial benign neonatal convulsions. Brain Dev 2009;31:27-33. [PubMed]
- Neubauer BA, Waldegger S, Heinzinger J, et al. KCNQ2 and KCNQ3 mutations contribute to different idiopathic epilepsy syndromes. Neurology 2008;71:177-83. [PubMed]
- Lange W, Geissendörfer J, Schenzer A, et al. Refinement of the binding site and mode of action of the anticonvulsant Retigabine on KCNQ K+ channels. Mol Pharmacol 2009;75:272-80. [PubMed]